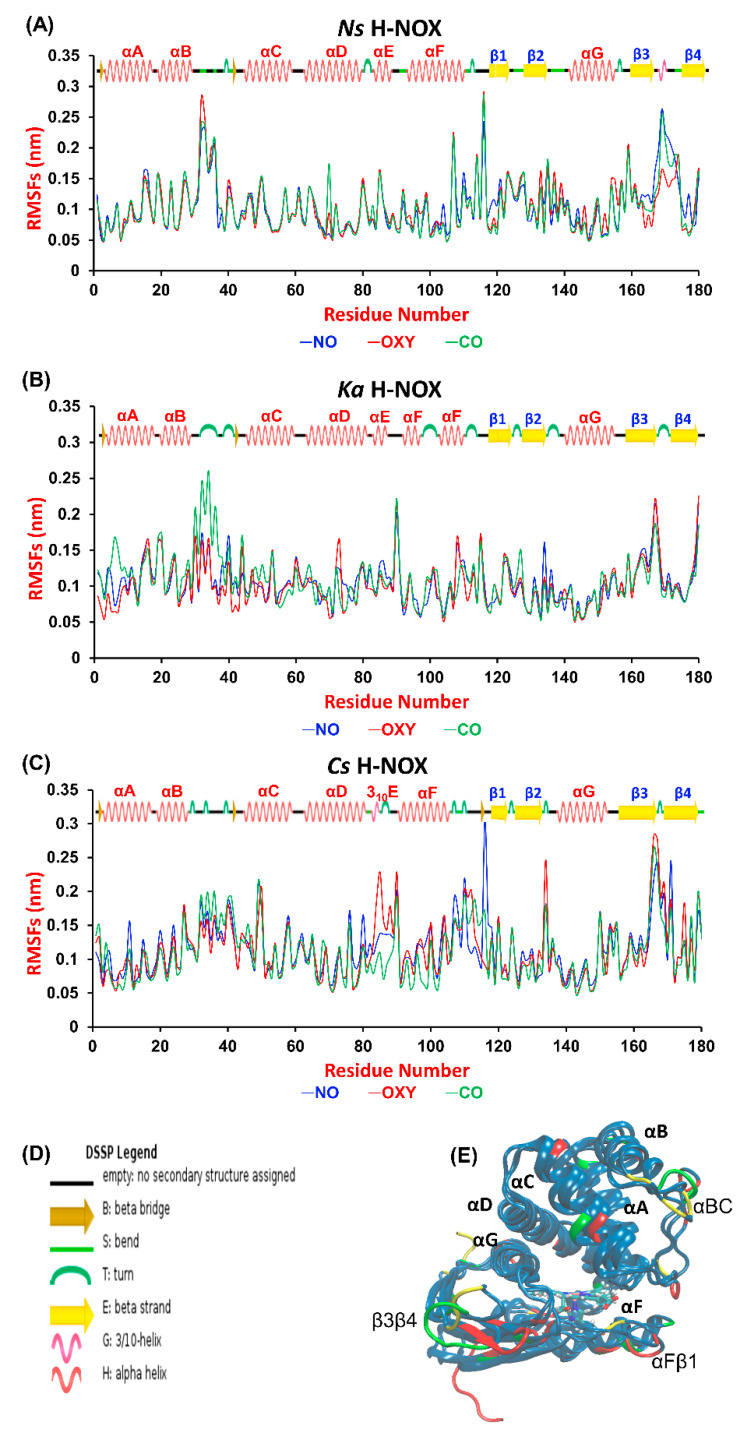
Figure 2.
RMSF values computed per-residue from molecular dynamics (MD) simulations of gaseous molecule (NO, CO, O2) diffusion into (A) Nostoc sp. (Ns), (B) Kordia algicida (Ka) and (C) Caldanaerobacter subterraneus (Cs) H-NOX. In the case of Cs, the RMSF values of the long tail (residues 181–188 that are not present in Ns and Ka H-NOX) are not shown to make comparison with other systems easier. (D) Labels used for the designation of secondary structure elements by DSPP in Figure 2A–C. (E) Superposition of the structure of the three H-NOX proteins. Regions with highest mobility have been highlighted by larger font size, and colored in green (Ns), yellow (Ka) and red (Cs).