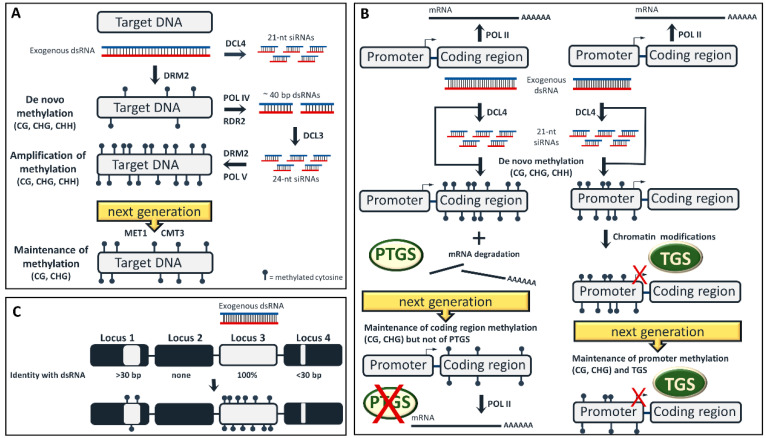
Figure 1.
Epigenetic modifications upon exogenous RNA interference (exo-RNAi) application. (A) The three steps of DNA methylation: de novo, amplification and maintenance. Exogenously applied double-stranded RNAs (dsRNAs) are mainly processed by dicer-like 4 (DCL4) into 21-nt short-interfering RNAs (siRNAs) that have no direct role in RNA-directed DNA methylation (RdDM) in the nucleus, but they mediate post-transcriptional gene silencing (PTGS) in the cytoplasm. According to our hypothesis, the exogenous long dsRNA triggers the first incomplete wave of de novo methylation, while the 24-nt siRNAs, that are subsequently generated upon RNA polymerase IV (POLIV) transcription of the de novo methylated locus, are seemingly involved in the amplification of these methylation marks. Thus, both long dsRNAs and 24-nt siRNAs are able to recruit domains rearranged methyltransferase 2 (DRM2) to their target in a stepwise manner. In the absence of RdDM trigger molecules (dsRNA or siRNAs) CHH methylation is mitotically/meiotically lost, while CG and CHG methylation are maintained by methyltransferase 1 (MET1) and chromomethylase 3 (CMT3), respectively. (B) Exogenous application of dsRNA designed to target a coding region (left) will result not only to mRNA degradation aPTGS, but also to RdDM of the coding region. In the next generation, PTGS will be lost, but DNA methylation (CG and to lesser extent CHG) will be maintained. Exogenous application of dsRNA designed to target a promoter (right) will lead to promoter RdDM. Should additional chromatin modifications occur, TGS will also take place. In the next generation, CHH methylation will be lost, but CG/CHG methylation and TGS will be maintained. (C) A dsRNA may trigger RdDM not only to a DNA sequence which shares full sequence identity (locus 3, on-target), but also to an unrelated DNA sequence exhibiting a minimum of 30-bp sequence identity with the dsRNA (locus 1, off-target).