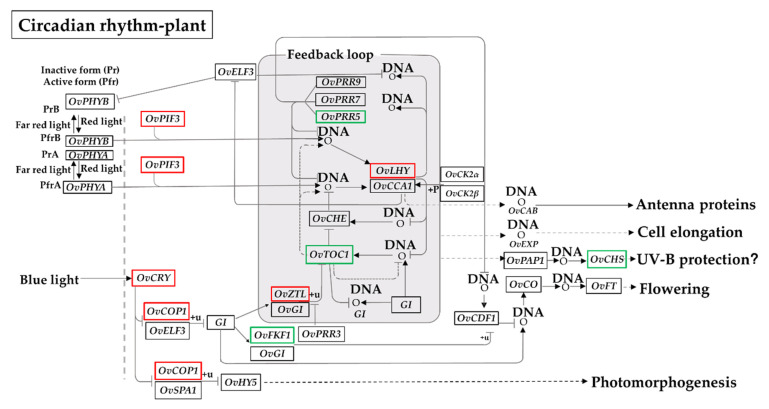
Figure 5.
A putative ‘circadian rhythm–plant’ pathway was generated from the KEGG database based on the log2 (FC) values of identified differentially expressed genes (DEGs). The symbols in boxes indicate the enzyme codes. Red and green colors depict the up- and down-regulated genes, respectively. OvPHYA, phytochrome A; OvPHYB, phytochrome B; OvELF3, EARLY FLOWERING 3; OvPIF3, phytochrome-interacting factor 3; OvPRR5, pseudo-response regulator 5; OvPRR7, pseudo-response regulator 7; OvPRR9, pseudo-response regulator 9; OvCRY, cryptochrome; OvCOP1, constitutive photomorphogenic 1; OvSPA1, suppressor of PHYA-105 1; OvGI, GIGANTEA; OvHY5, transcriptional factor HY5; OvCHE, transcription factor TCP21; OvZTL, ZEITLUPE; OvLHY, late elongated hypocotyl; OvCCA1, circadian clock associated 1; OvFKF1, flavin-binding kelch repeat F-box protein; OvCDF1, cycling dof factor 1; OvCHS, chalcone synthase; OvTOC1, timing of cab expression 1; OvCO, zinc finger protein CONSTANS; OvFT, FLOWERING LOCUS T; OvCK2α, casein kinase II subunit alpha; OvCK2β, casein kinase II subunit beta.