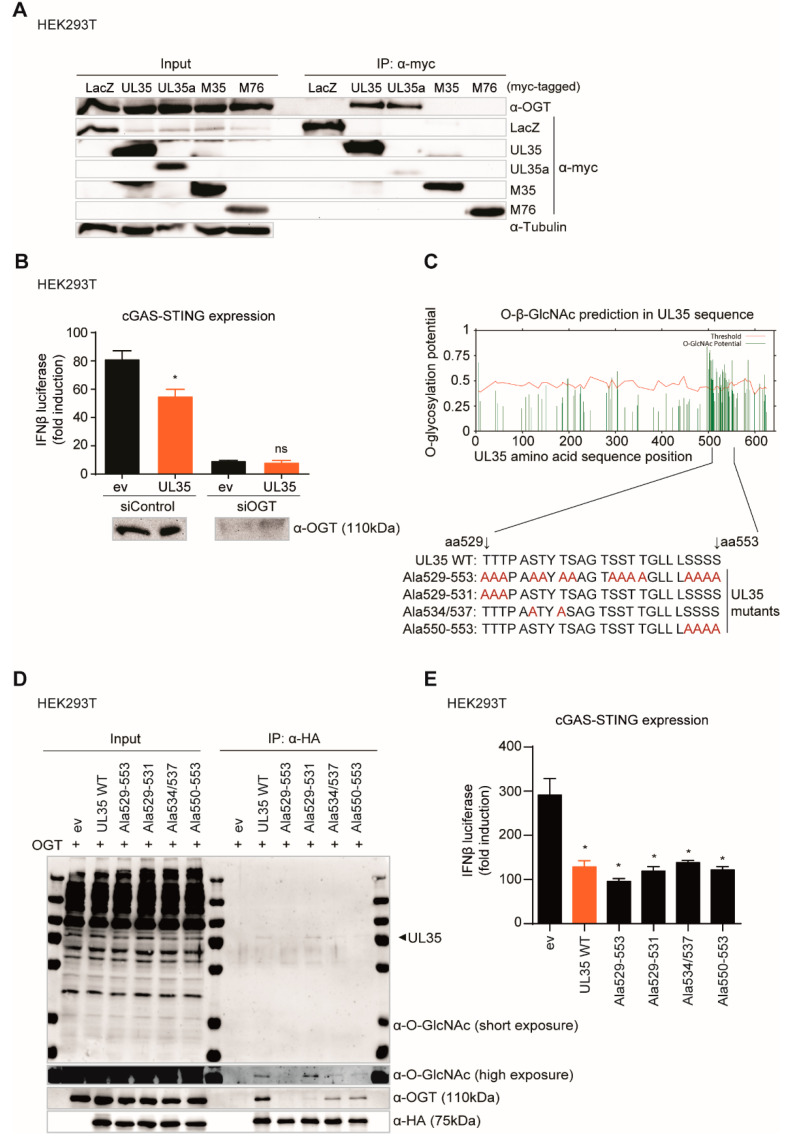
Figure 6.
GlcNAcylation and OGT interaction of UL35 is not needed for its antagonistic function. (A) HEK293T cells were transfected with myc-tagged LacZ, UL35, UL35a, M35, or M76 and cell lysates were subjected to immunoprecipitation (IP) with an anti-myc antibody. Input and IP samples were analyzed by immunoblotting with antibodies specific for OGT, myc, and tubulin. (B) HEK293T cells were reverse transfected with either control siRNA or siRNA specific for OGT. 48 h later, cells were co-transfected with cGAS (stimulated) or IRES-GFP (unstimulated), Cherry-STING, IFNβ-Luc, pRL-TK, and either ev or HA-tagged UL35. Lysates for measuring luciferase activity were collected 20 h later and analyzed as described before. (C) Potential O-GlcNAcylation sites were predicted within the UL35 amino acid sequence (Ref. seq. YP_081494.1) using the YinOYang 1.2 online server tool. High potential serine and threonine residues within UL35 aa positions 529–553 were mutated to alanines (depicted in red). (D) HEK293T cells were co-transfected with expression constructs for OGT and either ev or HA-tagged UL35 expression constructs. 20 h later, proteins were immunoprecipitated using an HA-specific antibody and separated by SDS-PAGE. GlcNAcylation was analyzed by immunoblotting with an anti-O-GlcNAc antibody. OGT interaction was verified with anti-OGT antibody and UL35 expression with an anti-HA antibody. (E) HEK293T cells were co-transfected with cGAS (stimulated) or IRES-GFP (unstimulated), Cherry-STING, IFNβ-luc, pRL-TK, and either ev or HA-tagged UL35 WT or UL35 alanine mutants. 20 h later, cells were lysed and luciferase activity was measured. (B,E) One representative experiment out of three independent experiments is shown as mean ± SD. Student’s t-test (unpaired, two tailed), UL35 was compared to ev. ns p > 0.05, * p < 0.05.