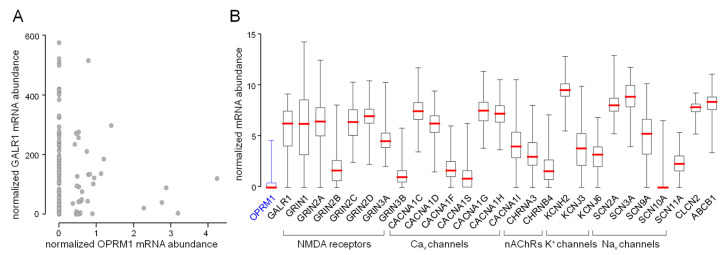
Figure 1.
Expression of methadone target molecules in glioblastoma (Illumina HiSeq_RNA Seq V2 glioblastoma dataset (n = 172) of The Cancer Genome Atlas (TCGA)). (A) Majority of glioblastoma specimens exhibit non-detectable low abundances of the µ-opioid receptor (OPRM1) mRNA. Here, the µ-opioid receptor mRNA abundances are plotted against those of the GALR1 (galanin receptor 1), which forms heteromeric receptors with OPRM1 (values of individual tumors are shown). (B) Box whisker plots depicting relative mRNA abundances (TCGA-normalized values) of proposed methadone targets in glioblastoma specimens (medians are highlighted by red lines). Cav channels: voltage-gated L- and T-type Ca2+ channels, nAChRs: nicotinic acetylcholine receptors, NMDA receptors: N-methyl-d-aspartate receptors, Nav channels: voltage-gated Na+ channels, KCNH2: hERG1 K+ channel, CLCN2: ClC-2 Cl− channel, ABCB1: p-glycoprotein (MDR1).