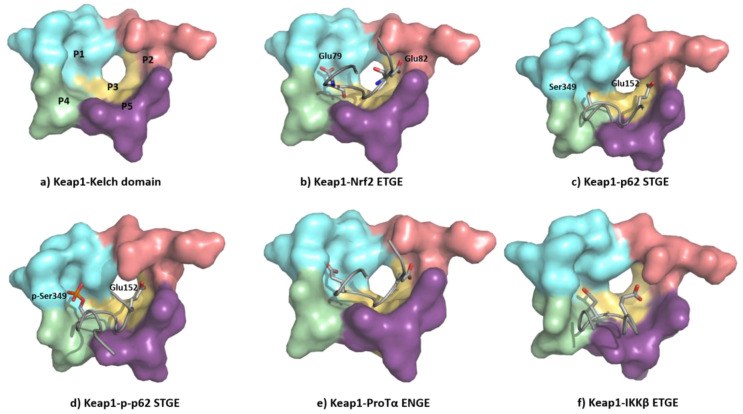
Figure 11.
(a) Structure of the Kelch domain of KEAP1 (PDB: 5WTV), highlighting binding pockets P1: blue (residues 415, 461, 462, 478, 483 and 508); P2: red (363, 380, 381 and 414); P3: yellow (364, 509, 556, 571, 602 and 603); P4: green (334, 572 and 577); P5: purple (525, 530 and 555); (b) structural detail of the NRF2 ETGE motif bound to KEAP1 (PDB: 5WTV); (c) structural detail of the p62 STGE motif (PDB: 3ADE). The STGE motif in the KEAP1-interacting region (KIR) is similar to the ETGE-like motif of NRF2 and they both adopt similar β-turn conformations. However, while Glu79 in ETGE interacts with the Arg415 contained in the KEAP1 P1 subpocket, the corresponding Ser349 in STGE is too short, leading to a poor binding affinity; (d) structural detail of the Ser349-phosphorylated p62 STGE motif (PDB: 3WDZ). Phosphorylated Ser439 is deeply inserted into the P1 subpocket, and the negative charges increase the affinity through electrostatic interactions with Arg415 and Arg483; (e) structural detail of Protα ENGE motif (PDB 2Z32); (f) structural detail of the IKKβ ETGE motif obtained by molecular dynamics [149]. The substitution of the Thr residue (DEETGE, NRF2) by an Asn residue (NEENGE, ProTα) renders the β-turn conformation unstable, leading to a less potent binding between the ENGE motif and KEAP1 compared with that of the ETGE motif in NRF2.