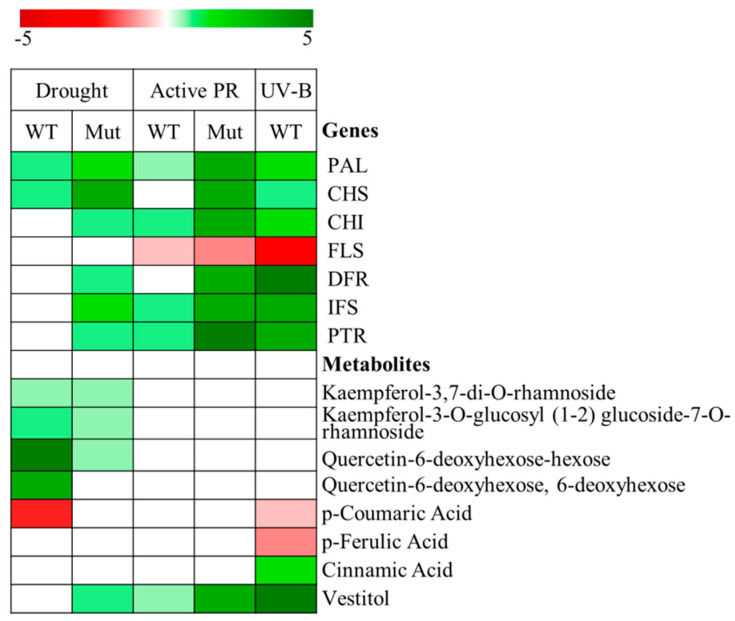
Figure 2.
Comparison of the changes in the relative expression levels of the most highly modulated genes (determined by RT-qPCR) and in the levels of metabolites in Lotus japonicus leaves, in response to different abiotic stress situations studied (drought, impairment of photorespiration, UV-B irradiation). Changes are indicated as log2 of the fold change in gene expression levels and metabolite content for each genotype and stress condition analyzed, relative to the unstressed control plants, as described in previous works [42,62]. Red and green indicate lower and higher levels than in the controls, respectively. The color intensity represents the log2 of fold change as indicated in the scale bar. WT, wild type; MUT, Ljgln2-2 photorespiratory mutant plants deficient in plastidic glutamine synthetase (GS2); active PR, active photorespiration. Active photorespiration is particularly stressful for the Ljgln2-2 mutant plants due to the high accumulation of ammonium produced as a consequence of the deficiency in photorespiratory ammonium re-assimilation (the impairment of photorespiration) [95,98]. Abbreviations used for genes are described in Figure 1. Given the high number of genes that encode for the flavonoid and isoflavonoid biosynthetic enzymes and the high gene redundancy, a set of specific oligonucleotides were utilized that amplified only specific copies of the redundant gene probesets for key enzymes of the pathways [62]. The results obtained for the most representative and highly expressed gene from each gene family are shown. Genes indicated have the following accession numbers (using Kazusa 3.0 terminology and when available GeneBank codes): PAL, Lj1g3v4590760 (BAF36971.1); CHS, Lj2g3v2124310; CHI, Lj5g3v2288880 (Q8H0G2.1); FLS, Lj1g3v0705350; DFR, Lj5g3v0108500 (BAE19948.1); IFS, Lj4g3v0485090 (BAF64284.1); PTR, Lj3g3v3360890 (BAF34841.1). Semi-systematic names and chemical structures of the indicated metabolites are shown in Table S1.