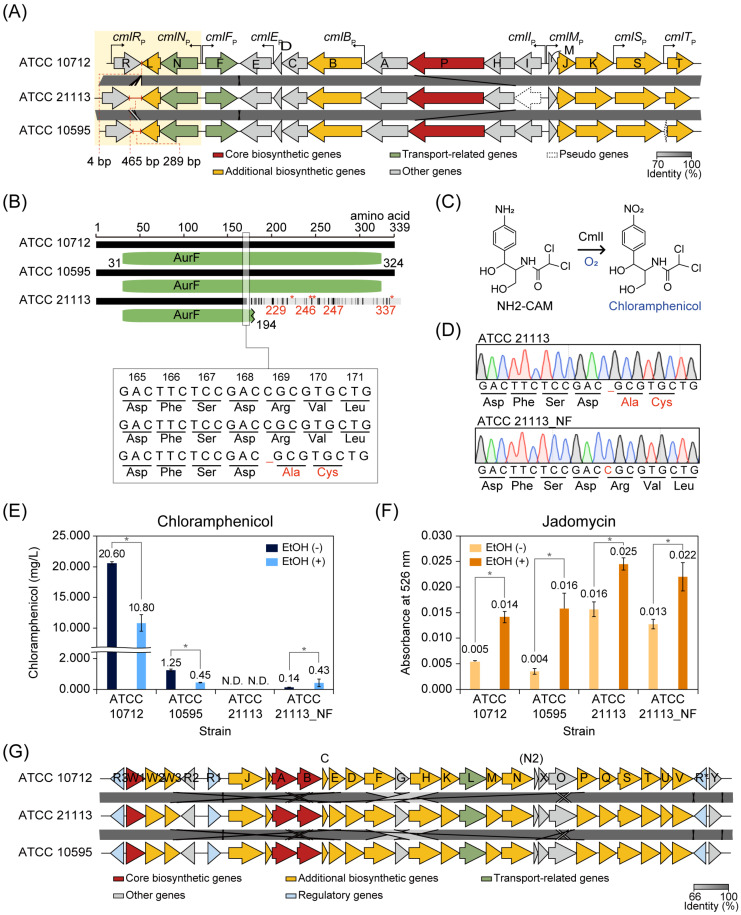
Figure 5.
Comparison of chloramphenicol and jadomycin production. (A) Alignment of the chloramphenicol biosynthetic gene cluster (BGC) from the three S. venezuelae strains. The region of cmlR and cmlLN operon was represented with a yellow box, and the red line indicates the intergenic region of cmlR and cmlL. The genes were colored according to the classification of antiSMASH prediction. Gray bars between the clusters indicate BLASTn identity value. The chloramphenicol biosynthetic genes were named, according to Sekurova et al. [50]. (B) Comparison of amino acid sequences of CmlI from the three S. venezuelae strains. Amino acid 31 to 324 of the CmlI was predicted as the AurF domain by Pfam [51], and it was represented as a green rounded rectangle. The asterisk indicates the position of a stop codon. (C) N-oxygenation of NH2-CAM by CmlI to produce chloramphenicol. (D) Sanger sequencing results of the cmlI gene from ATCC 21113 and ATCC 21113_NF. (E) Chloramphenicol production of the three S. venezuelae strains and ATCC 21113_NF in the absence or presence of ethanol. EtOH, ethanol; N.D., Not detected. Asterisk indicates p-value < 0.05. (Wilcoxon rank-sum test) (F) Jadomycin production of the three S. venezuelae strains and ATCC 21113_NF in the absence or presence of ethanol. Absorbance at 526 nm was measured for estimating jadomycin production. EtOH, ethanol; Asterisk indicates p-value < 0.05. (Wilcoxon rank-sum test) (G) Alignment of the jadomycin BGC from the three S. venezuelae strains. The genes were colored according to the prediction of antiSMASH. Gray bars between the clusters indicate BLASTn identity value. The jadomycin biosynthetic genes were named, according to Niu et al. [52].