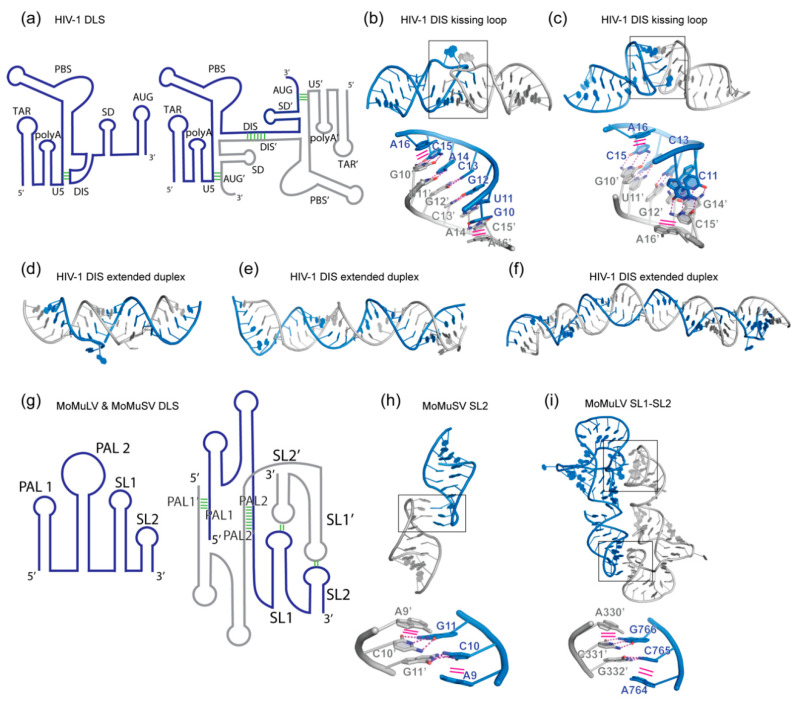
Figure 1.
Homodimerization of retroviral genomic RNAs (gRNA). (a) Schematic view of the 5′ leader dimer linkage site (DLS) of human immunodeficiency virus type 1 (HIV-1) driving gRNA dimerization. Left: monomeric state with the dimerization initiation site (DIS) sequestrated by the U5 segment. Right: dimeric state driven by intermolecular DIS-DIS’ interaction. The Gag start site AUG’ element pairs in trans with the U5 segment. These conformational changes allow the HIV-1 genome to switch between translation (monomeric) and packaging (dimeric) functions. Structural elements followed by a prime symbol indicate that they belong to the other protomer. (b,c) Crystal and NMR structure of the DIS kissing loop interaction, respectively (PDB 2B8S and 1BAU). (d,e,f) Crystal, NMR, and Cryo-EM structures of the DIS homodimer in its extended duplex state (PDB 1Y99, 2GM0 and 6BG9). (g) Schematic view of the 5′ leader structure of the Moloney murine leukemia virus (MoMuLV) and Moloney murine sarcoma virus (MoMuSV) regulating gRNA dimerization. Four elements in this RNA drive its dimerization: palindromic sequences in PAL1 and PAL2 and kissing loop interactions between Stem-loop 1 (SL1) and Stem-loop 2 (SL2). (h) Structure of MoMuLV homodimer SL2 (PDB 1F5U). (i) Structure of MoMuSV homodimer SL1-SL2 (PDB 2L1F). Interface regions are boxed and detailed below each structure. Hydrogen bonding interactions are shown as magenta dashed lines, while stacking interactions are shown as solid magenta lines.