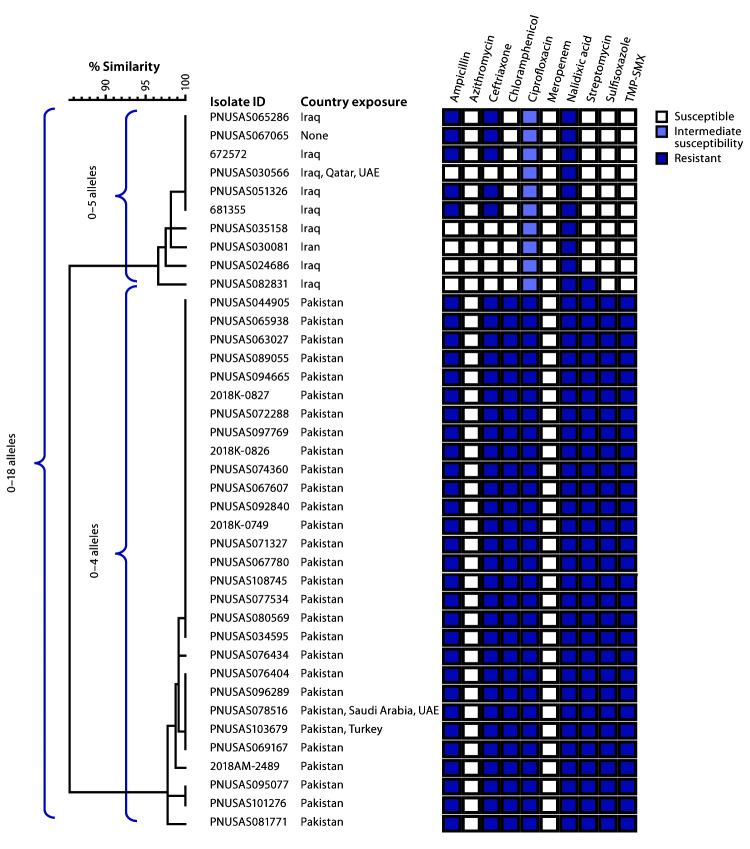
FIGURE 2.
Core genome multilocus sequence typing (cgMLST) phylogenetic tree* of 39† Salmonella Typhi isolates from two strains with ceftriaxone resistance among persons with travel to Iran, Iraq,§ and Pakistan — United States and United Kingdom,¶ 2017–2019.
Abbreviations: TMP-SMX = trimethoprim-sulfamethoxazole; UAE = United Arab Emirates.
* The tree was constructed using BioNumerics (version 7.6; Applied Maths). The National Center for Biotechnology Information strain identifier and country exposure(s) during the patient’s incubation period are shown. Shaded boxes indicate resistance patterns determined by antimicrobial susceptibility testing (n = 35) or predicted resistance from whole genome sequencing (n = 4, with isolate IDs 672572, 681355, PNUSAS095077, and PNUSAS101276).
† One ceftriaxone-resistant Typhi isolate from a patient with travel to Pakistan was not included in this figure because the isolate did not undergo whole genome sequencing.
§ One isolate (ID PNUSAS067065) was cultured from a patient who did not travel, but whose asymptomatic father returned from Iraq in the 30 days before her symptom onset.
¶ Isolates with IDs 672572 and 681355 were cultured from residents of the United Kingdom.