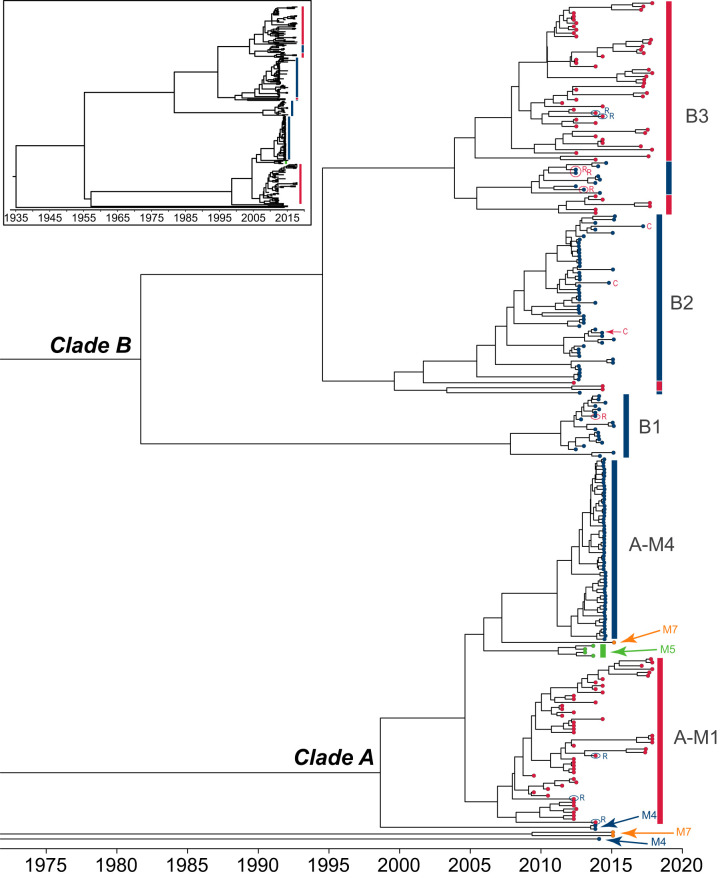
FIG 3.
Time-scaled phylogenetic reconstruction, showing a maximum clade credibility tree produced from the concatenated SNP alignment obtained with SLCC 2479 as the reference genome, using the Bayesian Markov chain Monte Carlo method available in BEAST (45). The inset shows the whole tree, while the main figure shows the section containing the tips, enlarged for clarity. Figure S4 shows the same tree, labeled with the posterior probability values for each split and node labels containing strain names. Isolates within cluster B2 were isolated from raw zone sampling points, except for the three nodes labeled with C (cooked zone). All other isolates were from cooked zones, except the five nodes labeled with circles and an R (raw zone). The tips of the branches correspond to the sampling date, while tip colors indicate factory of origin for each strain, colored as described for Fig. 1 and Fig. 2. The scale is in years.