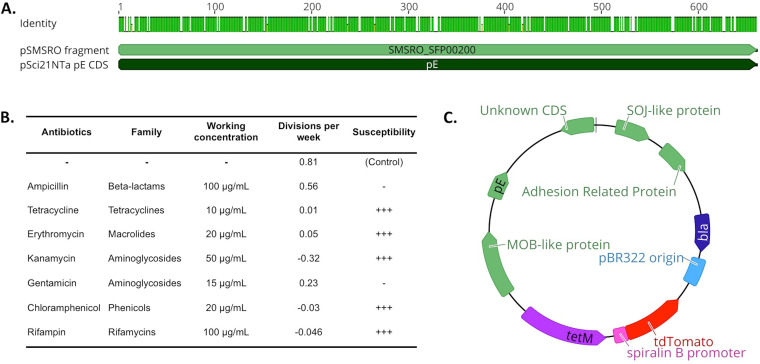
FIG 1.
Design of a fluorescence plasmid for S. poulsonii transformation. (A) Alignment of the sequence coding for the cis-acting plasmid replication protein pE in S. citri-derived plasmid pSci21NTa to the uncharacterized coding sequence SMSRO_SFP00200 located on S. poulsonii natural plasmid pSMSRO. A green mark on the “Identity” track indicates an identical amino acid for the considered position. The two sequences share 87% identity, suggesting that S. citri pE is likely to be functional in S. poulsonii. (B) Antibiogram of S. poulsonii in liquid culture. −, resistant; +++, susceptible. (C) Scheme of the pSpo-tdTomato plasmid. Blue indicates E. coli-optimized sequences for cloning, and green indicates S. citri coding sequences present on the original plasmid pSci2. Purple and red are Spiroplasma-optimized sequences. bla, beta-lactamase coding gene (ampicillin resistance marker for E. coli); tetM, gene encoding tetracycline resistance protein M (tetracycline resistance marker for Spiroplasma).