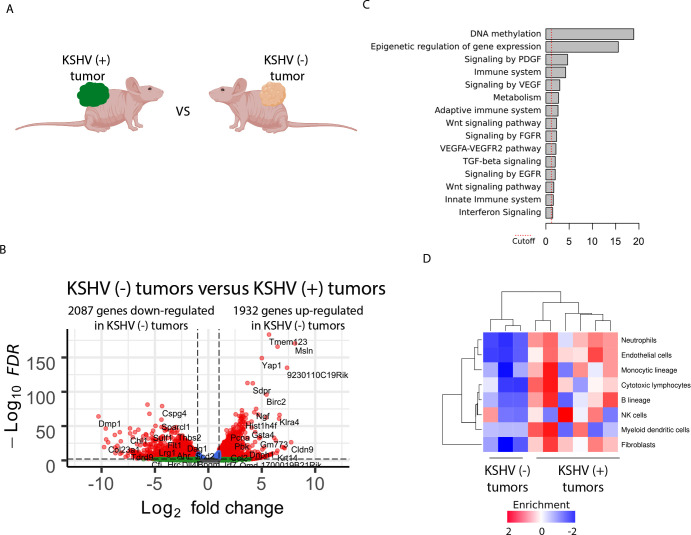
Fig 3. Transcriptional effects of KSHV infection in KSHV-positive and KSHV-negative tumors.
(A) Scheme of the comparison between KSHV (+) tumors and KSHV (-) tumors. (B) Volcano plot showing 4020 differentially expressed genes (DEGs) analyzed by RNA-Sequencing between KSHV (-) tumors and KSHV (+) tumors. 2087 genes were down-regulated and 1932 genes were up-regulated in KSHV (-) tumors. (C) Functional enrichment analysis based on the 4020 genes differentially expressed among KSHV (+) tumors and KSHV (-) tumors. (D) CIBERSORT in silico method to determine absolute immune cell fractions within the tumor microenvironment of KSHV-positive and KSHV-negative tumors.