Figure 2.
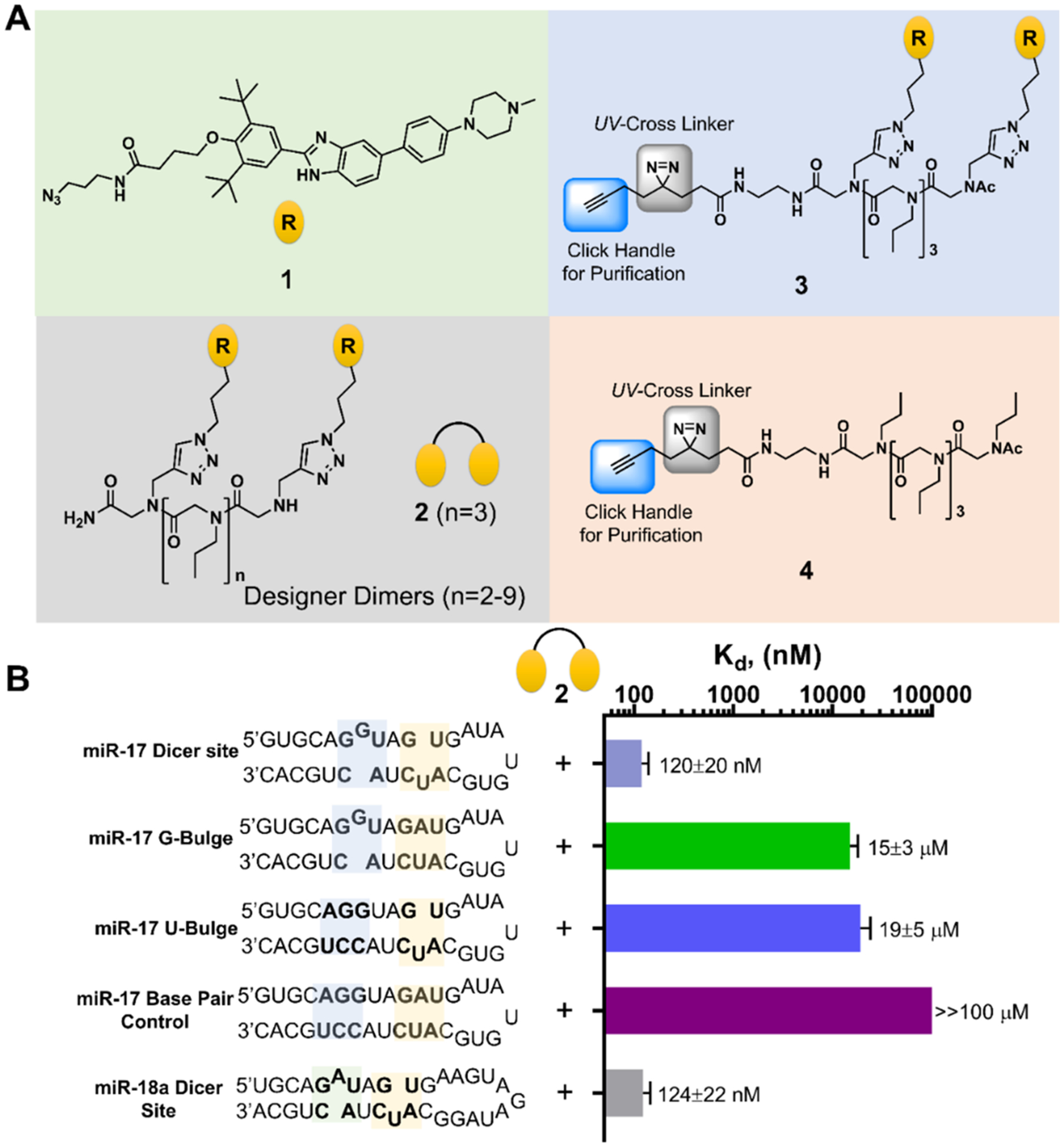
Design of a dimeric compound that binds to miR-17’s Dicer site. (A) Chemical structures of compounds used in these studies. Compound 1 is the parent monomer targeting each motif; 2 is the dimer with three propylamine spacing modules in the peptoid backbone, allowing specific binding to each miRNA’s Dicer site; 3 is a Chemical Cross-Linking and Isolation by Pull-down (Chem-CLIP) probe that contains a diazirine reactive module and click handle for conjugating biotin, enabling pull-down for assessing target engagement; 4 is the Chem-CLIP control probe that lacks the RNA-binding modules to assess nonspecific reaction of the diazirine. (B) Secondary structures of the model RNAs used in binding studies and the corresponding affinities for 2. Note that “miR-17 Dicer site” and “miR-18a Dicer site” are comprised of sequences native to the corresponding RNA. Other RNAs contain mutations that convert the bulges to base pairs.
