Figure 4. Fos/AP-1 Acts Downstream of EGFR to Stimulate Autophagy.
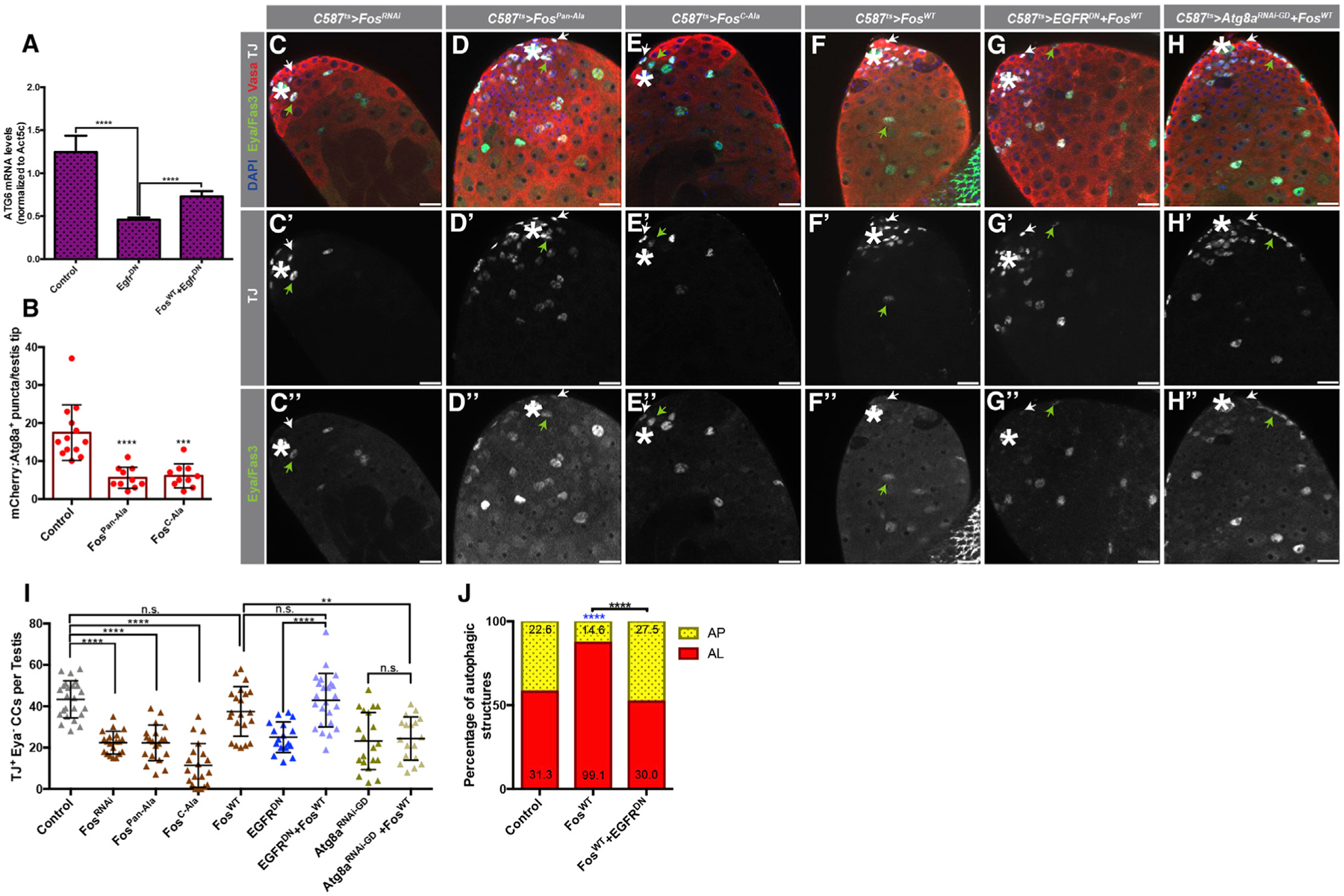
(A) qRT-PCR quantification of quintuplicates displaying the ratio between Atg6 and Atc5c mRNA in testes from 3-day-old animals of the noted genotypes.
(B) Quantification of the number of APs per testis tips in the somatic CC tissue of the noted genotypes.
(C–H’’) Representative images showing testes tips from 10-day-old animals expressing the early CC ‘driver’ c587-GAL4TS with: UAS-FosRNAi(C–C’’), UAS-FosPan-Ala(D–D’’), UAS-FosC-Ala(E–E’’), UAS-FosWT(F–F’’), UAS-EGFRDN, UAS-FosWT(G–G’’), and UAS-Atg8aRNAi-GD, UAS-FosWT(H–H’’). White arrows point to TJ+/Eya− CCs, and green arrows point to TJ+/Eya+ CCs. Note that all single UAS-based transgenes were paired with UAS-TdTomato to consider Gal4/UAS levels compared to double UAS combinations.
(I) Quantification of TJ+/Eya− CCs in 10-day-old animals of the noted genotypes. Control data reproduced from Figure 3J for comparison.
(J) Quantification of autophagic structures in the noted genotypes. Blue asterisks represent statistical information compared to control. Numbers on each barcolor represent the mean number of autophagic structures per testis tip (n = 10 testes per condition). Control data reproduced from Figure 1F for comparison. In all images, asterisks (*) denote the hub; scale bars, 20 μm. For (A), (B), and (I), two-tailed t tests were used; error bars represent standard deviation. For (J), two-sided Fisher’s exact was used. *p < 0.05, **p < 0.01, ***p > 0.001, ****p < 0.0001; n.s., not significant.
