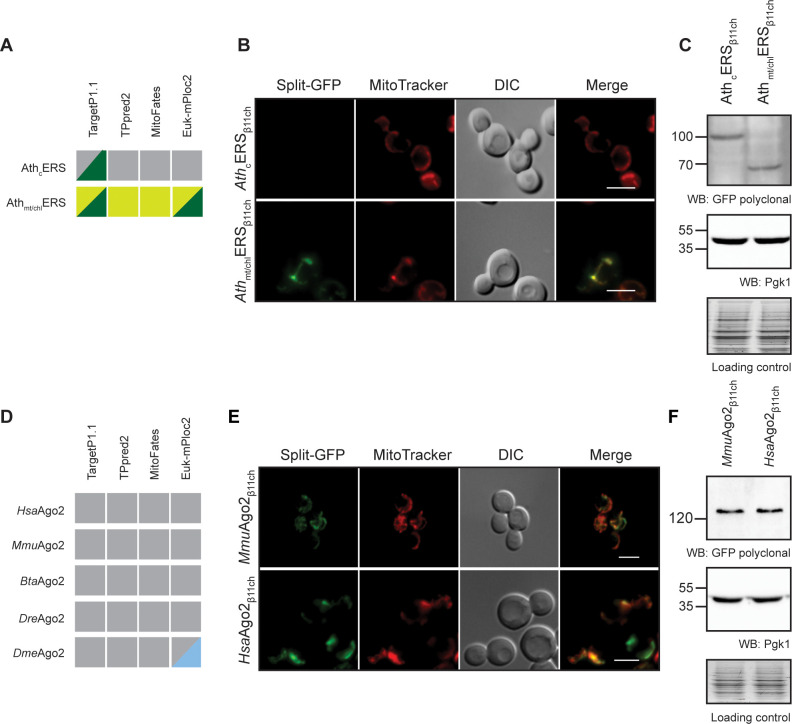
Figure 5. The BiG Mito-Split-GFP can be used to study mitochondrial importability of mammalian and plant proteins.
(A, D) Prediction of MTS and mitochondrial localization of (A) two ERS from Arabidopsis thaliana (AthcERS and Athmt/chlERS) and (D) five eukaryotic Ago2 proteins [HsaAgo2 (Protein argonaute-2 isoform X2 [Homo sapiens] NCBI sequence ID: XP_011515267.1), MmuAgo2 (protein argonaute-2 Mus musculus NCBI sequence ID: NP_694818.3.), BtaAgo2 (Bos Taurus), DreAgo2 (Danio rerio), DmeAgo2 (Drosophila melanogaster). MTS were predicted using TPpred2.0 (http://tppred2.biocomp.unibo.it/tppred2), TargetP1.1 (http://cbs.dtu.dk/services/TargetP/), MitoFates (http://mitf.cbrc.jp/MitoFates/cgibin/top.cgi) and the EukmPloc2 website (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/). Grey boxes indicate prediction of a cytosolic localization, light and dark green indicate prediction of mitochondrial or chloroplastic localization respectively. Blue boxes indicate prediction of nuclear localization. (B, E) Fluorescence microscopy analyses of the BiG Mito-Split-GFP strain expressing the GFPβ11ch-tagged AthcERS and Athmt/chlERS (N = 2) (B) andMmuAgo2, HsaAgo2 (N = 2) (E). Mitochondria were stained with MitoTracker Red CMXRos. Scale bar: 5 µm. Representative fields are shown. (C, E) Protein expression was checked by WB with anti-GFP antibodies and equal amount of loaded protein was controlled using anti-Pgk1 antibodies and by the stain-free technology (Loading control: stain-free). The representative gels are shown.