Figure 2: Overview of the SingleCellExperiment class.
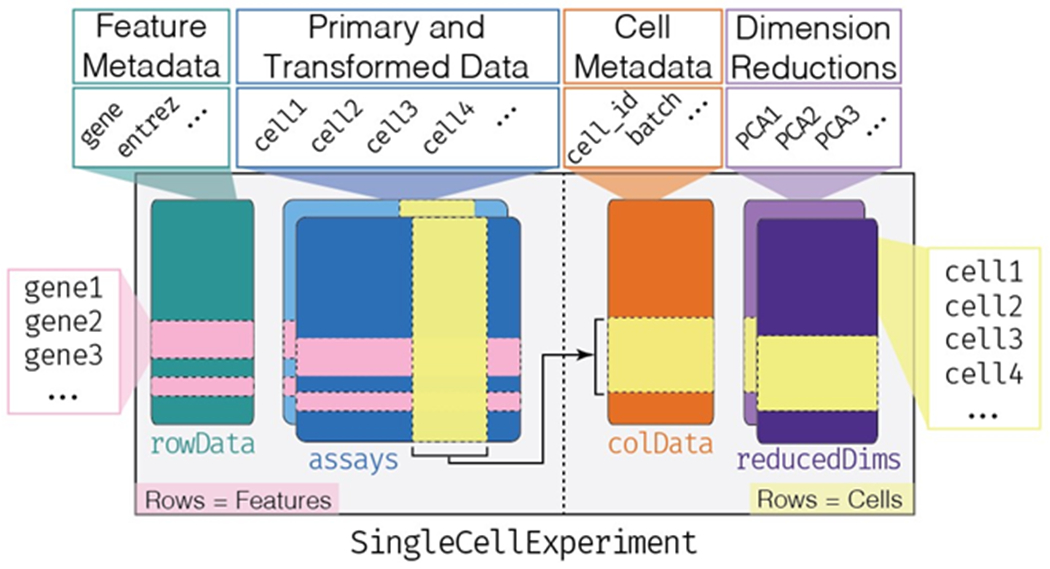
The SingleCellExperiment class instantiates an object (SingleCellExperiment herein abbreviated sce) capable of storing various datatypes associated with single-cell assays. A sce object is organized into components (e.g. rowData, assays, colData, reducedDims). In the assays component the rows represent features such as genes (horizontal pink bands), and the columns represent cells (vertical yellow band). The rowData and colData components can hold information (such as metadata) about the features and cells, respectively. Note that in the colData and reducedDims components, cells are represented as rows (horizontal yellow bands) and the number of columns in the assays component must match the number of rows in the colData and reducedDims components.
