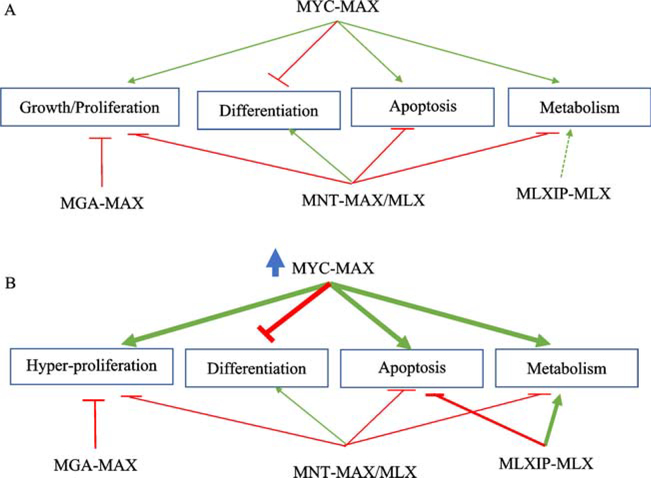
Fig. 5.
A hypothetical representation of two states of the MYC network and their impact on gene expression programs that influence growth, proliferation, differentiation, apoptosis and metabolism. (A) A balanced network in which gene expression is controlled through normal endogenous regulation of the MYC network. Transcriptional effects of MYC-MAX are balanced by MNT (heterodimerized with either MAX or MLX), MGA-MAX, and, under conditions of stress, by MLXIP-MLX or MLXIPL-MLX. (B) An unbalanced network due to deregulation of MYC expression. In this state MYC-MAX suppresses differentiation, reprograms metabolism, and triggers the apoptotic pathway. The suppressive effects of MGA-MAX and MNT-MAX/MLX on proliferation are overwhelmed by MYC-MAX which contributes to suppression of MYC-induced apoptosis. Increased nuclear accumulation of MLXIP-MLX (in response to deregulated MYC and/or metabolic stress) adjusts metabolic reprogramming by MYC-MAX and further reduces apoptosis. The effects on gene expression are presumed to occur through genomic binding and co-occupancy by network members. Green arrows, transcriptional activation; red arrows, transcriptional repression. Arrow width proportional to estimated transcriptional effect. Diagram adapted from [5].