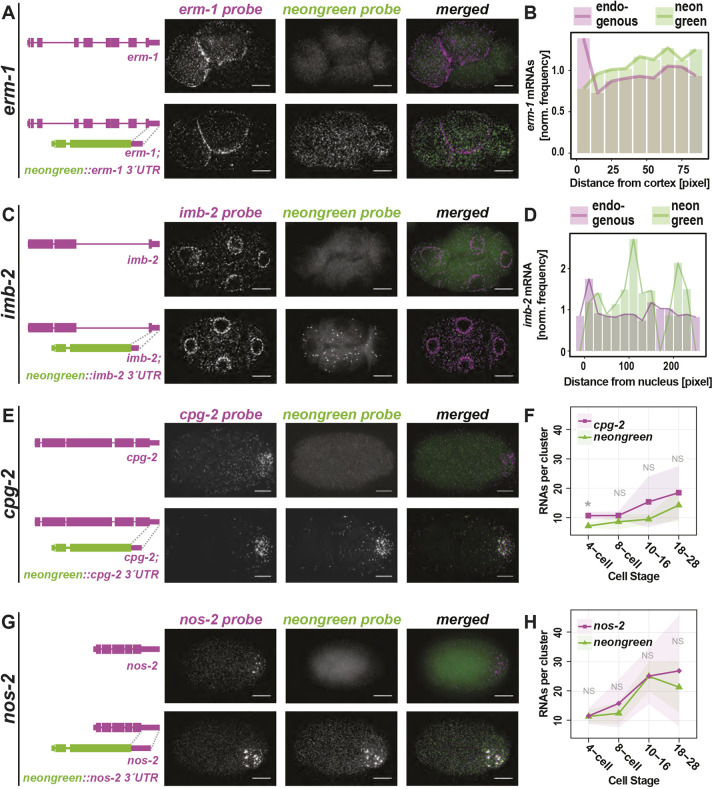
Fig. 4.
3′UTRs of clustered, but not membrane-associated, transcripts are sufficient for subcellular localization. (A,C,E,G) The 3′UTRs of erm-1 (A), imb-2 (C), cpg-2 (E) and nos-2 (G) were appended to monomeric NeonGreen (mex-5p::mNeonGreen::3′UTR of interest) and transgenically introduced as a single copy insert into otherwise wild-type worms. Wild-type control strains (top panels) and transgenic strains (bottom panels) were imaged by smFISH using probes hybridizing to the endogenous mRNA of interest (left) and to mNeonGreen mRNA (middle) and merged (right). Representative four-cell stage embryos are shown. (B,D) Quantification of images shown in A and C indicating the normalized frequency of erm-1 (B) or imb-2 (D) mRNA and mNeonGreen mRNA at increasing distances from cell peripheries or nuclear boundaries, respectively, in a single embryo. (F,H) The estimated mRNA content per cluster from a minimum of five embryos at each of five binned stages of development from three biological replicates are reported for endogenous cpg-2 (F) or nos-2 (H) (magenta) and mNeonGreen reporters (green). P-values from multiple test corrected t-tests are shown (NS>0.05; 0.05>*>0.005). Scale bars: 10 µm.