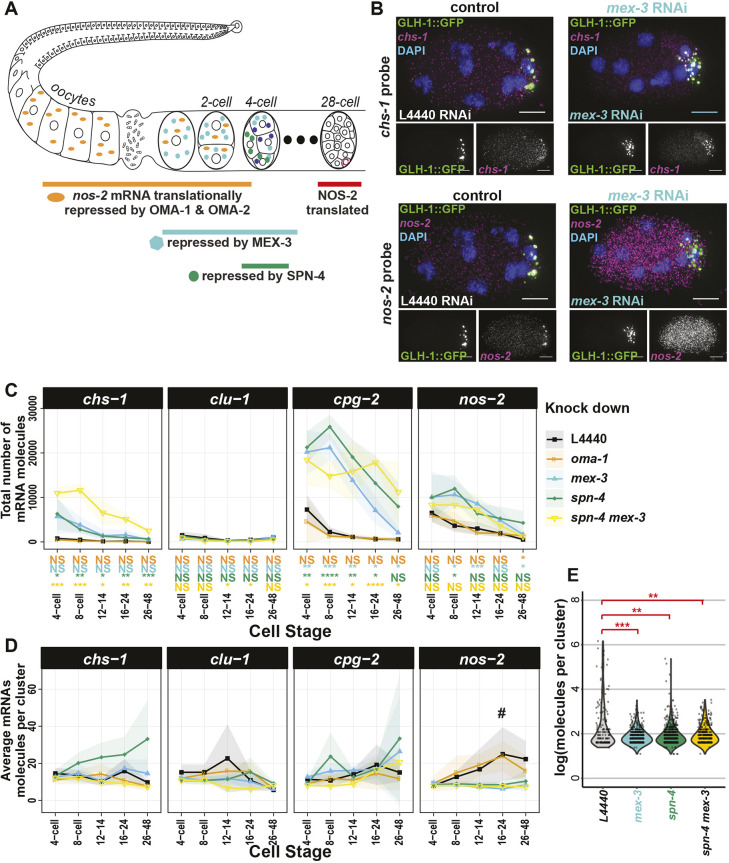
Fig. 5.
RBPs that repress translation of nos-2 mRNA also impact degradation rates and subcellular localization of key mRNAs. (A) A succession of RBPs cooperatively repress nos-2 translation from oogenesis through to the 28-cell stage. (B) chs-1 mRNA (magenta, top) and nos-2 mRNA (magenta, bottom) were imaged by smFISH in a P granule marker strain (GLH-1::GFP, green) under mock (L4440) and mex-3 RNAi conditions. (C,D) The total number of mRNA molecules (C) and average number of mRNA molecules per cluster (D) for four different RBP knockdown conditions on five mRNA at five different developmental stages are shown graphically, compared with the L4440 empty vector RNAi control. At least four embryos were assayed for each data point from three biological replicates. Standard deviations are shown as shaded ribbon regions. # indicates data analyzed in E. (E) Distributions of nos-2 mRNA cluster size under mex-3, spn-4 (ts), and dual mex-3/spn-4 depletion conditions at the 16- to 24-cell stage demonstrate decreased cluster sizes when compared with mock (L4440) depletion. Significance indicates P-values derived from multiple test corrected t-tests comparing the knockdown condition of interest with vector-only RNAi control (L4440) (0.005>**>0.0005; 0.0005>***>0.00005; 0.00005>****). Scale bars: 10 µm.