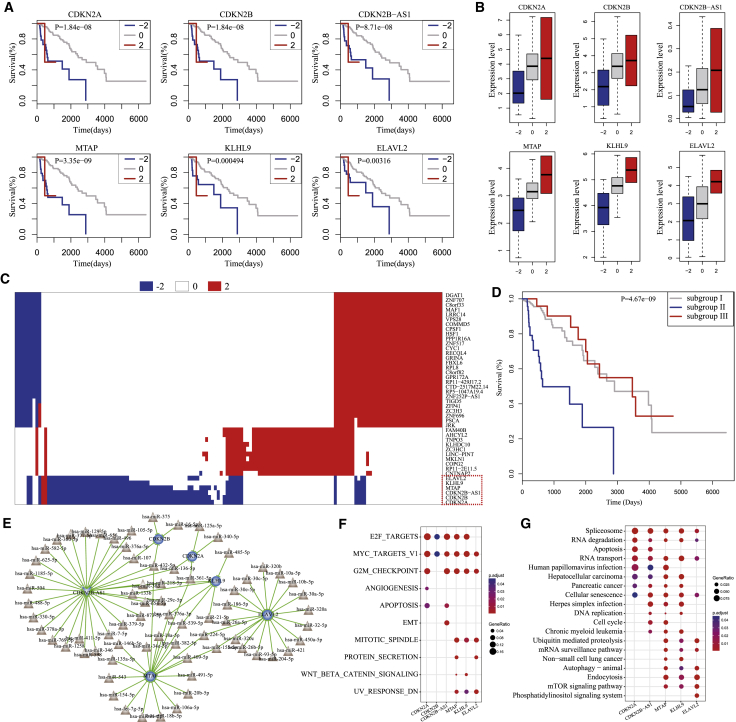
Figure 4.
The Homozygous Deletion of 9p21.3 Characterized a LGG Subtype with Poor Prognosis
(A) The homozygous deletions of six genes, including CDKN2B-AS, CDKN2B, CDKN2A, MTAP, KLHL9, and ELAVL2 were associated with poor prognosis in LGG. (B) The homozygous deletions of six genes significantly decreased their own gene expression. (C) The heatmap for SCNAs of 44 driver genes across LGG patients. (D) 9p21.3 deletions characterized a new LGG subtype with poor prognosis. (E) Comprehensive skeleton of the dysregulated ceRNA networks induced by homozygous deletions of six driver genes. (F) Hallmark signatures significantly enriched by the six dysregulated ceRNA networks. (G) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways significantly enriched by the six dysregulated ceRNA networks.