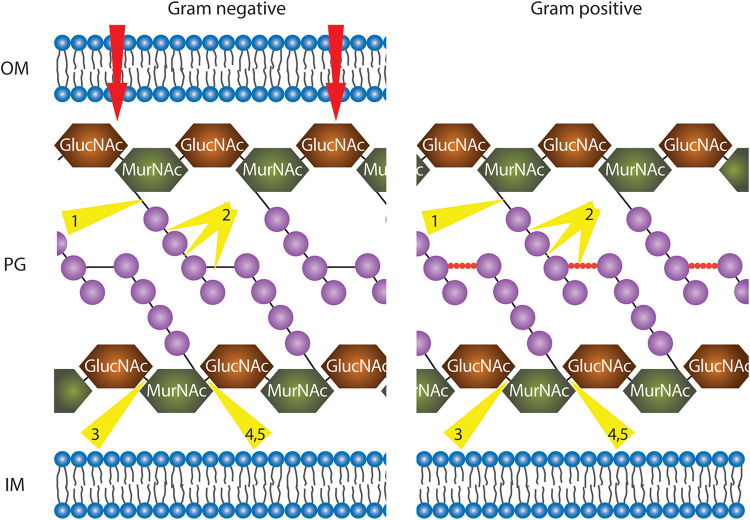
FIGURE 4.
Illustration of phage endolysins classified by their enzymatic activity. In order to reach the peptidoglycan (PG) layer, in the case of Gram-negative bacteria, the outer membrane (OM) has to be traversed (red arrows, details in text). The PG layer is represented with alternate repeating sugar units, N-acetylglucosamine (GlucNAc), and N-acetylmuramic acid (MurNAc). In Gram-negative bacteria, tetrapeptide chains (purple spheres) extend from each MurNAc of the long sugar chains and are directly cross-linked by short interpeptide bridges (black solid line) via the third residue to D-Ala of another peptide chain. In Gram-positive bacteria, the tetrapeptide chains (purples spheres) are linked via pentapeptide bridges (red spheres). All residues are purple spheres. IM, inner membrane. Depending on the organism, residues at position 3 are either L603 Lysine (mainly Gram-positive bacteria) or meso-diaminopimelic acid (Gram-negative bacteria and Gram-positive bacilli). Endolysin cleavage sites are indicated by yellow triangles: 1, N-acetylmuramoyl-L-alanine amidases; 2, endopeptidases; 3, endo-N-acetyl606 glucosaminidases; 4, N-acetyl muramidases; 5, lytic transglycosylases.