Figure 2.
KLF3 Is Necessary for Human Epidermal Differentiation
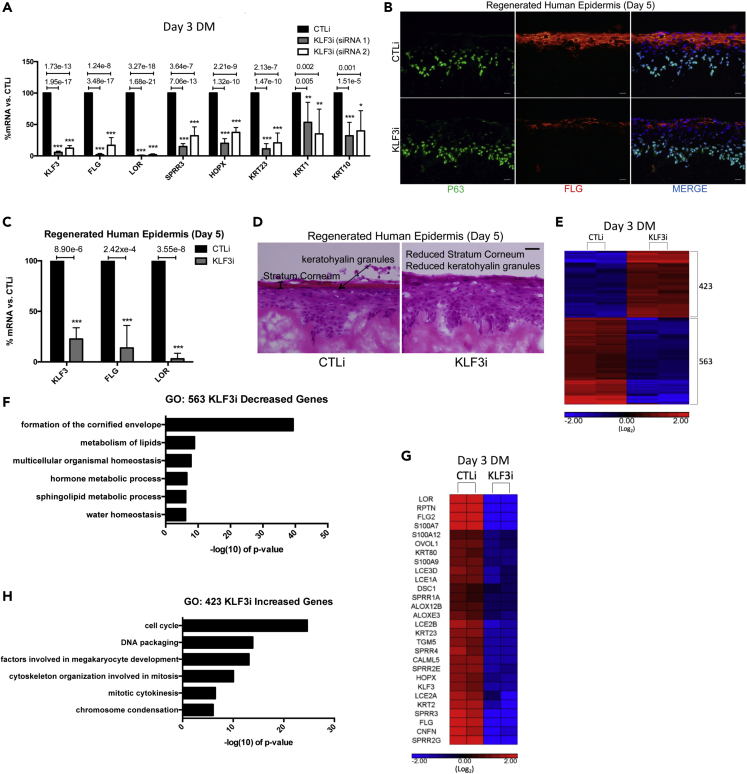
(A) RT-qPCR quantifying the relative mRNA expression levels of a panel of epidermal differentiation genes in CTLi and KLF3i keratinocytes after 3 days of differentiation. Two separate siRNAs (siRNA 1 and siRNA 2) targeting different regions of KLF3 were used (n = 6). Data are graphed as the mean ± SD. Statistics: t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(B) Immunofluorescent staining of late differentiation marker FLG (red) and basal layer marker P63 (green) in day 5 regenerated human epidermis treated with control (CTLi) or KLF3 targeting (KLF3i) siRNA. Merged image includes Hoechst staining of nuclei. n = 3. Scale bar: 20 μm.
(C) RT-qPCR quantifying the relative mRNA expression levels of LOR and FLG in CTLi and KLF3i day 5 regenerated human epidermis. n = 4. Data are graphed as the mean ± SD. Statistics: t test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(D) Hematoxylin and eosin staining of CTLi and KLF3i day 5 regenerated human epidermis. n = 3, scale bar: 20 μm.
(E) Heatmap generated for replicate (n = 2) RPKM normalized RNA-Sequencing data from CTLi and KLF3i keratinocytes differentiated for 3 days. The expression of genes significantly (FDR ≤0.05 and fold change ≥2 versus CTLi) increased (red) or decreased (blue) are displayed (log2 scale).
(F) Gene ontology (GO) term enrichment for the 563 genes significantly decreased upon KLF3 knockdown.
(G) Heatmap showing the relative expression levels of a panel of differentiation-associated genes from the CTLi and KLF3i RNA sequencing datasets (log2 scale). n = 2.
(H) Gene ontology term enrichment for the 423 genes significantly increased in expression upon KLF3 knockdown.