Figure 6.
KLF3 Is Necessary for CBP Localization at Enhancers Proximal to Differentiation Genes
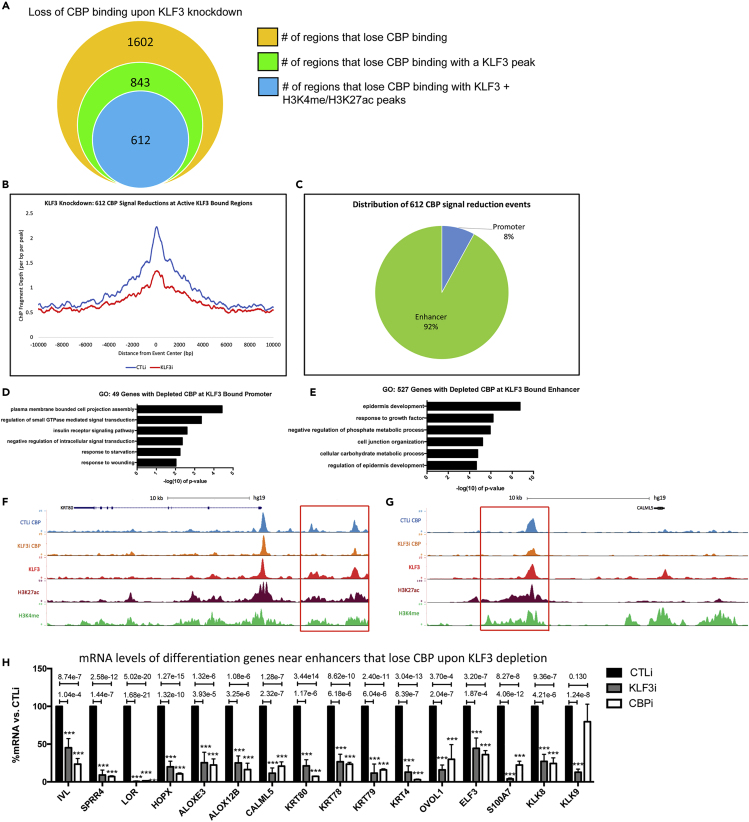
(A) Summary of significant CBP ChIP signal reduction events in day 3 differentiated keratinocytes knocked down for KLF3. The orange circle represents the total number of regions in the genome that lose CBP binding upon KLF3 knockdown. The green circle represents the number of regions that lose CBP binding that occur at a KLF3 peak upon KLF3 depletion. The blue circle represents the regions that lose CBP binding, which also contain KLF3, H3K4me, and H3K27ac binding upon KLF3 knockdown. CBP ChIP Seq was performed in replicates in CTLi and KLF3i cells. Significant signal changes were identified by Diffreps.
(B) Mean density profile displaying CBP binding centered around KLF3/H3K4me/H3K27ac peaks. The CBP mean density is plotted around the 612 regions identified in (6A:blue circle). The profile of CBP binding is shown +/− 10kb from the centers of the 612 regions in CTLi (blue) and KLF3i (red) cells.
(C) Percent distribution of the 612 H3K27ac/H3K4me positive regions containing KLF3 peaks with significant CBP depletion upon KLF3 knockdown. These 612 regions were annotated using HOMER and all regions not mapped to promoters were considered enhancers.
(D) Gene ontology (GO) term enrichment for the 49 genes that lose CBP binding at their active promoter (H3K4me/H3K27ac positive) containing a KLF3 peak.
(E) Gene ontology term enrichment for the 527 genes that lose CBP binding at a proximal enhancer (H3K27ac/H3K4me positive) containing a KLF3 peak. (F) UCSC genome browser tracks displaying CBP binding (CBP ChIP Seq profiles) from CTLi (blue) and KLF3i (orange) cells, near differentiation gene KRT80. KLF3 (red), H3K27ac (maroon), and H3K4me (green) binding are also shown.
(G) UCSC genome browser tracks displaying CBP binding (CBP ChIP Seq profiles) from CTLi (blue) and KLF3i (orange) cells, near differentiation gene CALML5. KLF3 (red), H3K27ac (maroon), and H3K4me (green) binding are also shown.
(H) RT-qPCR quantifying the relative mRNA expression levels of epidermal differentiation genes near enhancers that lose CBP genomic binding upon KLF3 knockdown in CTLi, KLF3i, and CBPi keratinocytes after 3 days of differentiation. N = 4 except for LOR and HOPX in KLF3i samples where N = 6. Statistics: t test, ∗∗∗p < 0.001. Mean values are shown with error bars = SD.