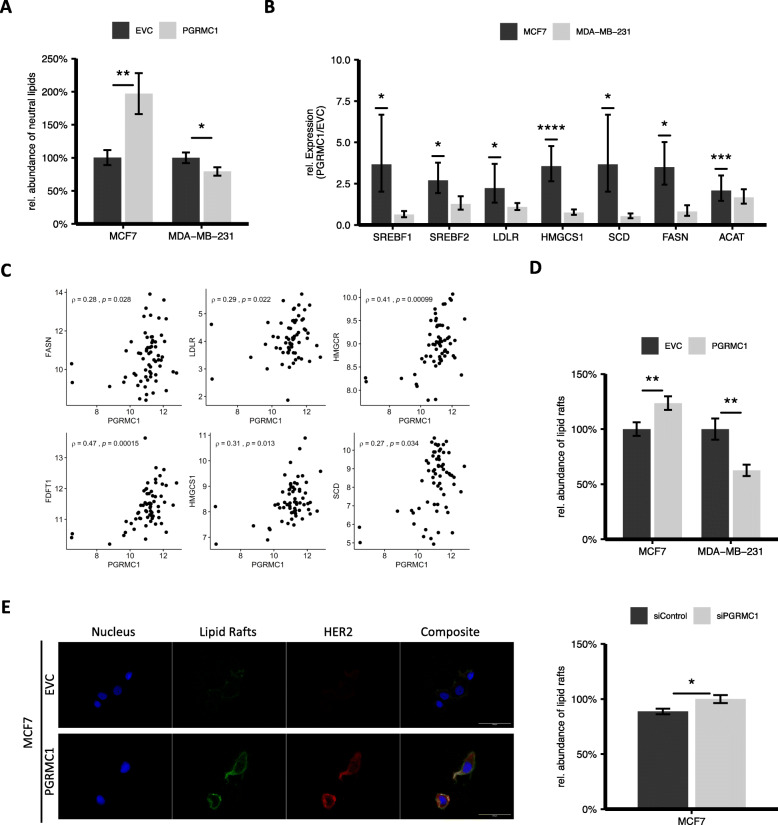
Fig. 4.
a Detection of neutral lipids and lipid droplets in MCF7/EVC and MCF7/PGRMC1, MDA-MB-231/EVC and MDA-MB-231/PGRMC1 cells by BODIPY® staining and quantification via flow cytometry. *p ≤ 0.05, **p ≤ 0.01. (Student’s t test, n = 3). b qRT-PCR analysis of SREBF1, SREBF2, LDLR, HMGS1, SCD, FASN, ACAT mRNA expression in MCF7/PGRMC1 and MDA-MB-231/PGRMC1 cells compared to the respective EVC cells. *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 (Student’s t test, n = 3). c Spearman’s correlation between the PGMRC1 expression level and various expression levels of proteins (FASN, FDFT1, HMGCS1, HMGCR, LDLR, SCD) involved in lipid metabolism. Data obtained from normalized microarray data (Affymetrix Human Genome U133A Array) of 63 hormone receptor-positive breast cancer tissue samples. d Detection of lipid rafts in cell membranes of MCF7/EVC and MCF7/PGRMC1 cells, MDA-MB-231/EVC and MDA-MB-231/PGRMC1 cells, and MCF7 siCtrl and MCF7 siPGRMC1 cells by Vybrant™ Alexa Fluor™ 488 and subsequent quantification via flow cytometry. *p ≤ 0.05, **p ≤ 0.01 (Student’s t test, n = 3). e Immunofluorescence staining with Vybrant™ Alexa Fluor™ 488, fluorescence immunocytochemistry for HER2, and nuclear staining with DAPI. 63-fold magnification. Cells were grown on chamber slides for 24 h