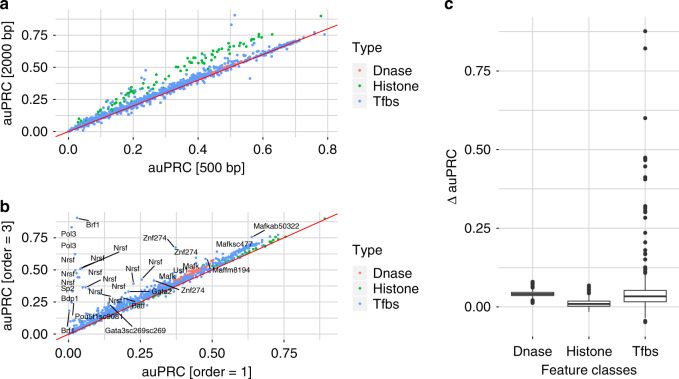
Fig. 3. Comparison of DanQ model variants.
a auPRC comparison for the context window sizes 500 bp and 2000 bp for tri-nucleotide based sequence encoding. The mark color indicates the feature types: DNase hypersensitive sites, histone modifications and transcription factor binding assays. b auPRC comparison for tri- and mono-nucleotide based sequence encoding for a context window of 2000 bp. Color coding as above. c Differences in auPRC between tri- and mono-nucleotides for DNase accessibility, histone modifications and transcription factor binding, respectively. ΔauPRC > 0 indicates improved performance for the tri-nucleotide based encoding. Dnase, histone modification and TF features comprise n = 125, n = 104, and n = 690 samples, respectively. Boxes represent quartiles Q1 (25% quantile), Q2 (median), and Q3 (75% quantile); whiskers comprise data points that are within 1.5 x IQR (inter-quartile region) of the boxes.