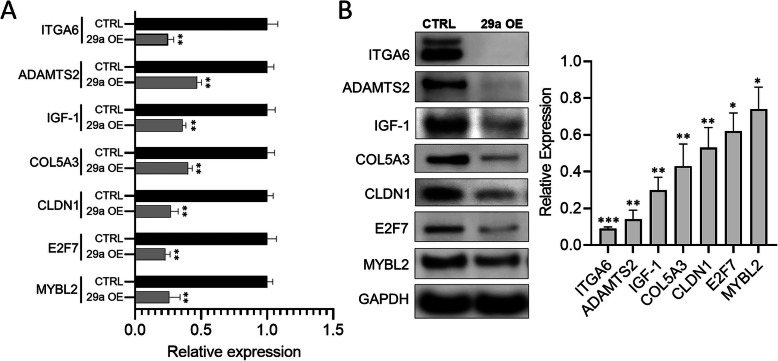
Fig. 2.
Validation of miR-29a direct target. a Relative fold changes estimated by qPCR analysis for the top miR-29a candidate target genes of ITGA6, ADAMTS2, IGF-1, COL5A3, CLDN1, E2F7 and MYBL2 in hPSCs transfected with miR-29a mimics (29a OE) compared with cells transfected with scramble control (CTRL). Numerical data are represented as average fold change (ΔΔCT) ± standard error of the mean (SEM); **p < 0.01; n = 3. b Total protein harvested from the hPSCs transfected with scramble control (CTRL) or miR-29a mimics (29a OE) 48 h post-transfection were subjected to western blot analysis for miR-29a candidate targets of ITGA6, ADAMTS2, IGF-1, COL5A3, CLDN1, E2F7 and MYBL2. GAPDH was used as the loading control. Quantification of band intensities normalized to GAPDH. Quantification of band intensities normalized to GAPDH and relative to respective controls are represented as ± SEM; n = 3, *p < 0.05, **p < 0.01, ***p< 0.001 (right). Uncropped blots are shown in Additional file 3: Fig. S1