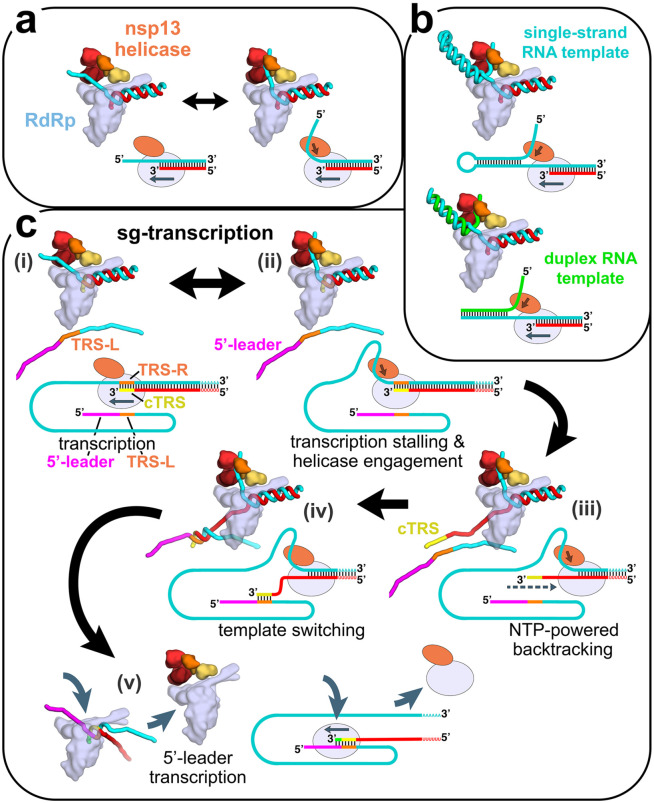
Figure 6. Structural basis for possible nsp13 helicase functions during viral genome replication/transcription.
Structural models are shown as cartoons (holo-RdRp, light blue; nsp13.1 helicase, orange shades; RNA strands, colored tubes). The nsp13.2 helicase is not shown for clarity (all of the models are compatible with the presence of nsp13.2). With each structural diagram is a schematic cartoon illustrating the arrangement of RNA strands. Additional proteins involved in these processes are omitted. The product RNA (p-RNA) being elongated by the RdRp is shown in red.
a. The SARS-CoV-2 nsp13-RdRp cryo-EM structure likely represents an equilibrium between two states.
b. During RNA synthesis on a single-stranded RNA template (cyan), nsp13 could function distributively to clear downstream RNA secondary structure (or RNA binding proteins). Similarly, on a duplex RNA template (cyan and green), nsp13 could processively unwind downstream duplex RNA.
c. Proposed helicase function during template-switching associated with sub-genomic (sg) transcription [see (Enjuanes et al., 2006; Lehmann et al., 2015b; Pasternak et al., 2001; Snijder et al., 2016; Sola et al., 2015)].
i) Negative-strand RNA synthesis proceeds from the genomic 3’-poly(A)-tail until a Transcription-Regulating Sequence [TRS-R, orange; (Alonso et al., 2002)] is transcribed (cTRS, yellow).
ii) The TRS causes transcription complex stalling.
iii) Helicase function acting on the +-strand RNA (cyan) causes backtracking of the transcription complex, freeing the pRNA 3’-end.
iv) The p-RNA 3’-end cTRS (yellow) hybridizes with the complementary TRS-L (orange) following the genomic 5’-leader sequence [magenta; (Alonso et al., 2002; Pasternak et al., 2001; Zúñiga et al., 2004)].
v) Processive helicase function backtracks the RdRp complex and unwinds the p-RNA from the genomic 3’-end. A second RdRp complex can load into the p-RNA 3’-end and continue transcription using the 5’-leader as template.