Figure 3.
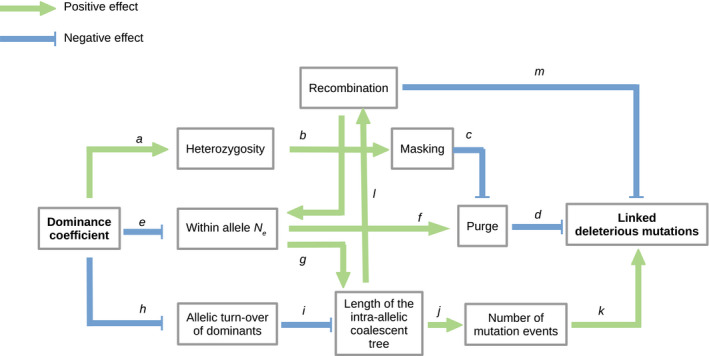
Theoretical predictions on the link between dominance and the load of deleterious mutations (see section 3.3). The formation of homozygous for the same S‐haplotype requires a cross between two heterozygotes sharing the same recessive and not expressed S‐allele. The formation of homozygous is thus impossible for the most dominant class. b. An increase in heterozygosity for an S‐haplotype also reduces the expression of recessive deleterious mutations associated with this haplotype. Yet, the recessive alleles occur more often at homozygous state so that their associated deleterious mutations are more often expressed. c. The purge of recessive deleterious alleles associated with S‐haplotype in the population is accelerated by their expression in the homozygous state. The latter is reduced when the dominance coefficient increases, leading to an increase in the genetic burden associated with dominant alleles; d. The stronger the purge is, the fewer deleterious mutations linked to an S‐haplotype are expected. e. In case of total co‐dominance between S‐alleles, the population is expected to be isoplethic for allele frequencies. With hierarchical dominance relationships, recessive S‐alleles can segregate at higher frequencies than the dominant ones, with a larger effective allele size than the dominant ones. f. For a slightly deleterious mutation linked to an S‐haplotype, the increase in the Ne.s product (where Ne is the effective population size and s corresponds to the effect of the mutation on individual fitness) will increase the efficiency of the purging of this mutation from the population. g. The total length of the intra‐S‐haplotype coalescing shaft increases for high Ne values. An increased tree length will increase the population mutation rate theta (j), and the population recombination rate rho (l). h. Recessive S‐haplotypes are more likely to be lost by drift as compared to dominant ones that immediately benefit from negative frequency‐dependent selection when they occur at rare frequency within populations. i. Faster turnover of the recessive alleles reduces the average age of alleles compared to dominant alleles that segregate in the population over longer evolutionary times. k. A greater number of mutations in the coalescent tree are associated with an increase in the number of deleterious mutations. m. Recombination breaks the correlation in gene genealogies between the selected target (S‐locus) and the linked regions
