Figure 4.
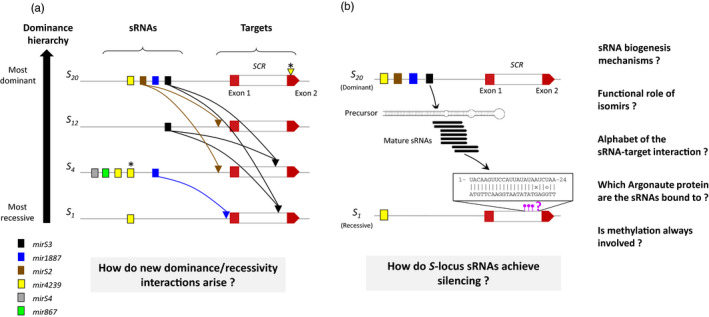
Dominance hierarchy among SI alleles in Arabidopsis. (a) Dominance can be explained by the evolution of a complex of sRNA regulatory network (see section 4.2). Schematic representation of the molecular regulatory network for four A. halleri S‐locus haplotypes chosen to be representative of the sRNA/target interactions identified in Durand et al. (2014), and ordered according to their dominance hierarchy. The repertoire of sRNA precursor families is represented by colored boxes, and SCR alleles are displayed for each haplotype. Target predictions, based on the sequence similarity between sRNAs and SCR alleles for all pairwise combinations, are displayed by curved arrows pointing to the target position (promoter or intron) when they are consistent with the phenotype, or marked by a star if not (here, only one predicted interaction opposed to the dominance phenotype was detected between S4 mir4239 and exon2 in SCR2 0). (b) Open questions on the mechanistic control of dominance (see section 4.4). Focus on one interaction to illustrate how dominance modifiers achieve silencing of the recessive alleles. The mirS3 precursor of the dominant haplotype S20 (represented by a black rectangle) is transcribed and produces a short hairpin structure, which is processed into mature sRNAs which recognize the intron of SCR of the recessive haplotype S1 based on high sequence similarity between the mature sRNA and the target
