Abstract
Efforts for sharing individual clinical data are gaining momentum due to a heightened recognition that integrated data sets can catalyze biomedical discoveries and drug development. Among the benefits are the fact that data sharing can help generate and investigate new research hypothesis beyond those explored in the original study. Despite several accomplishments establishing public systems and guidance for data sharing in clinical trials, this practice is not the norm. Among the reasons are ethical challenges, such as privacy of individuals, data ownership, and control. This paper creates awareness of the potential benefits and challenges of sharing individual clinical data, how to overcome these challenges, and how as a clinical pharmacology community we can shape future directions in this field.
Data are the building blocks of information that are critical for the modern advancement of human research. Over the past decade, we have witnessed rapid advances in technology, which led to the daily generation of an unprecedented amount of data and the development of platforms that can facilitate storing and sharing of these data.1, 2 Today, sharing data and knowledge are critical for the advancement of any research field. In clinical trial research, an enormous amount of data are generated at every stage of the clinical trial cycle, which is crucial for the drug development process. However, not sharing these data could undercut many benefits.3 In this review paper, we will discuss the potential benefits and expected challenges of sharing individual‐level clinical data, how to overcome these challenges, and how as a clinical pharmacology community we can shape future directions in this field.
Potential Benefits of Clinical Data Sharing
Sharing clinical data holds the promise of improving reproducibility and transparency of clinical research by allowing researchers to validate one another’s findings and decrease the impact of publication bias.4 Additionally, it could open opportunities for researchers, scientists, and drug authorities to analyze and translate clinical trial data to gain additional knowledge and strengthen the evidence for regulatory and clinical decisions.5 Responsible sharing of clinical trials data could also help in generating new research hypotheses about the safety and efficacy of drug therapies and investigating new questions and analytical methods beyond those planned in the original study.6 It could also avoid the duplication of effort in data collection, reduce unnecessary costs of future studies, and encourage collaboration and data circulation within the scientific community to inform decision making for clinical research planning and policy.7, 8 Moreover, freeing clinical data could help researchers, clinicians, and scientists to build upon each other’s work by conducting well‐powered meta‐analysis to have fast, robust, and more meaningful conclusions of the benefits and risks of a therapeutic intervention.9 These strong conclusions would empower healthcare professionals to make informed clinical care decisions and improve public health and patients’ outcomes by producing better evidence on the efficacy and safety of drugs.10, 11
Because of all the potential benefits of sharing data (Figure 1), over the past 2 decades, several efforts have been performed to establish public systems and guidance for data sharing in clinical trials, as discussed in the following section.
Figure 1.

Benefits of clinical trial data sharing.
Data Sharing in Clinical Trials
Throughout the last 2 decades, great accomplishments have been stridden toward establishing public systems and guidance for data sharing in clinical trials (Figure 2). In 2000, a publicly open web‐based database, ClinicalTrials.gov, was launched by the US Food and Drug Administration (FDA) Modernization Act (FDAMA) of 1997.12 In 2007, registration of clinical trial protocol summary became a requirement to submit to ClinicalTrials.gov by the FDA Amendments Act (FDAAA) regulations.13 These systems enable the public to access clinical trial findings with a comprehensive search.14, 15
Figure 2.
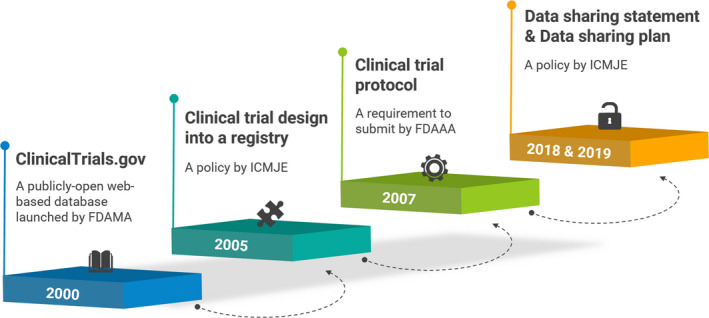
Accomplishments of establishing public systems and guidance for data sharing in clinical trials. FDAAA, Food and Drug Administration Amendments Act; FDAMA, Food and Drug Administration Modernization Act; ICMJE, International Committee of Medical Journal Editors.
These accomplishments also took place for journal publications. The International Committee of Medical Journal Editors (ICMJE) defined a clinical trial as “a research project that prospectively assigns human subjects to intervention or comparison groups to study the cause‐and‐effect relationship between a medical intervention and a health outcome” in 2005. In the same year, ICMJE initiated a policy requiring authors to register their trial design into a clinical trial registry, which is accessible to the public and electronically searchable.16, 17 As of July 1, 2018, ICMJE requires authors to include a data‐sharing statement in manuscripts presenting any findings from clinical trials.18 In addition, submitting a data‐sharing plan and reporting of any changes after registration became requirements for clinical trials that begin enrolling participants as of January 1, 2019.18 The requirements are expected to reduce any existence of publication bias.15, 19
These inspirational systemically achievements toward end‐to‐end data sharing in clinical trials are also supported by clinical trial participants.20 Among 421 completed surveys from diverse clinical trials, > 80% of respondents were willing to share their data with scientists working in for‐profit companies as well as academic research institutions. Only a few responders (< 8%) showed concern that the potential risks of data sharing could outweigh its benefits. This study laid to rest the mistrust and concerns that sharing data could make people hesitate to participate in clinical trials.
Approaches for Clinical Data Sharing
If routine clinical data sharing is to become a requirement, the approaches to support data sharing must be well‐defined. Sharing of individual‐level data can be performed mainly via a minimal or an expanded approach.21, 22 A minimal approach can be one in which a researcher or a small group of researchers does responsible sharing of de‐identified individual‐level data online in an isolated manner. Although with merits, this approach makes it difficult for potential users to become aware of the data, understand the data structure and specifications, integrate the data with other studies, and use the data in a proper context. On the other hand, an expanded approach can be one in which an entity disseminates standardized and anonymized clinical data to a broader research community with a mandate. The data sharing would be governed by data contribution and use agreements and could occur through an online data repository. Such a repository could include data specifications, tools for data querying, and some means of support. Examples of expanded data‐sharing initiatives are presented in Table 1.
Table 1.
Examples of expanded data‐sharing initiatives (original source reference49
| Initiative | Focus area | Description | Outcomes | URL |
|---|---|---|---|---|
| ADNI | AD and MCI | A longitudinal study that aims to identify clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of AD | 1,736 peer‐reviewed publications to date | http://adni.loni.usc.edu/ |
| AIBL | AD and MCI | A longitudinal study to determine which biomarkers, cognitive characteristics, and health and lifestyle factors determine subsequent development of symptomatic AD | 333 peer‐reviewed publications to date | http://adni.loni.usc.edu/aibl‐australian‐imaging‐biomarkers‐and‐lifestyle‐study‐of‐ageing‐18‐month‐data‐now‐released/ |
| CPAD Consortium | AD and MCI | CDISC standardized integrated clinical trial database of placebo arms in AD from 28 clinical trials contributed by industry partners | Alzheimer’s Disease Clinical Trial Simulation Tool endorsed by the FDA and the EMA, predementia clinical trial enrichment tool letter of support from the EMA | https://c‐path.org/programs/cpad/ |
| dbGaP | Interaction of genotypes and phenotypes for various diseases | Data‐sharing platform to archive and distribute the data and results from studies that have investigated the interaction of genotype and phenotype in humans | > 3,000 peer‐reviewed publications and abstracts | https://www.ncbi.nlm.nih.gov/gap/ |
| Enroll‐HD | HD | A worldwide observational study for HD families designed to accelerate the discovery and development of new therapeutics for HD | > 300 open and completed projects to accelerate HD research in disease progression, drug discovery, and preclinical/clinical research | https://www.enroll‐hd.org/learn/about‐this‐study/ |
| ImmPort | Allergy, Autoimmune diseases, Infection responses, Transplantation, and Vaccine responses | An open‐access data repository of subject‐level human immunology data, with a commitment to promoting effective data sharing across the basic, clinical, and translational research communities | 391 studies including 116 clinical trials shared to date; 385 peer‐reviewed publications cited ImmPort for data sharing, tools, data reuse, and secondary analysis | https://www.immport.org/shared/home |
| MSOAC | MS | CDISC standardized integrated clinical trial database of placebo arms in MS from nine clinical trials contributed by industry partners | EMA draft qualification opinion on a test battery for MS | https://www.ema.europa.eu/en/documents/scientific‐guideline/draft‐qualification‐opinion‐multiple‐sclerosis‐clinical‐outcome‐assessment‐mscoa_en.pdf |
| PPMI | PD | Observational clinical study to verify progression markers in PD | 131 peer‐reviewed publications to date | http://www.ppmi‐info.org/ |
| PKD Consortium | PKD | CDISC standardized database consisting of de‐identified data from three longitudinal observational patient registries for PKD | Total kidney volume as a prognostic biomarker for PKD endorsed by the FDA and the EMA | https://c‐path.org/programs/pkd/ |
| PDS | Oncology | A digital library‐laboratory that provides one place where the research community can broadly share, integrate, and analyze historical, patient‐level data from academic and industry phase III cancer clinical trials | 150 peer‐reviewed publications to date | https://projectdatasphere.org/projectdatasphere/html/home |
| ReseqTB data platform | TB | A data platform that catalogs genotypic, phenotypic, and related metadata from mycobacterium TB strains to enable the development of clinically useful, WHO‐endorsed in vitro diagnostic assays for rapid drug susceptibility testing of the bacteria | 14 peer‐reviewed publications to date | https://platform.reseqtb.org/ |
| TB‐TB‐PACTS | TB | CDISC standardized integrated clinical trial database of placebo arms in TB from 17 phase III clinical phase | A comprehensive pooled analysis of data from the database guided an optimal clinical trial design by helping to quantify the types of patient populations needed for optimal treatment regimes50 | https://c‐path.org/programs/tb‐pacts/ |
| WWARN | Malaria | A global platform that provides research evidence to support international efforts to fight antimalarial drug resistance | 123 peer‐reviewed publications to date | https://www.wwarn.org/ |
| National Cancer Institute GDC | Oncology | Data‐sharing platform that promotes sharing of genomic and clinical data between researchers to facilitate precision medicine in oncology | 95 peer‐reviewed publications to date | https://gdc.cancer.gov/ |
| TCIA | Oncology | An open‐access data repository of medical images for cancer patients to promote sharing of oncology clinical imaging data and advance our understanding of cancer |
> 750 peer‐reviewed publications to date |
https://www.cancerimagingarchive.net/ |
AD, Alzheimer's Disease; ADNI, Alzheimer's Disease Neuroimaging Initiative; AIBL, Australian Imaging, Biomarker and Lifestyle Flagship Study of Ageing; CDISC, Clinical Data Interchange Standards Consortium; CPAD, Critical Path for Alzheimer's Disease; dbGaP, Database of Genotypes and Phenotypes; EMA, European Medicines Agency; FDA, US Food and Drug Administration; GDC, Genomic Data Commons; HD, Huntington’s disease; ImmPort, Immunology Database and Analysis Portal; MCI, mild cognitive impairment; MS, Multiple sclerosis; MSOAC, Multiple Sclerosis Outcome Assessments Consortium; PDS, Project Data Sphere; PKD, Polycystic Kidney Disease; PPMI, Parkinson’s Progression Markers Initiative; ReseqTB, Relational Sequencing TB Data Platform; TB, tuberculosis; TB‐PACTS, TB‐Platform for Aggregation of Clinical TB Studies; TCIA, The Cancer Imaging Archive; WHO, World Health Organization; WWARN, WorldWide Antimalarial Resistance Network.
Although some of the expanded data‐sharing initiatives include data that have been collected under a single (e.g., Parkinson’s Progression Markers Initiative) or similar research protocols (e.g., Alzheimer’s Disease Neuroimaging Initiative (ADNI)), others integrate data from various clinical studies, hence, collected under different protocols (e.g., Critical Path Institute consortia). The latter requires a series of stages, as presented in Figure 3. An expanded data‐sharing initiative should be fit‐for‐purpose, intended to address an unmet medical need yet with specific research questions, and involve collaboration among stakeholders. This would constitute the foundation for the pursuit of clinical study data sets, which would be then transferred in a de‐identified and anonymized format23 through a secure link after the Data Contribution Agreement had been signed. In addition to the minimal or expanded approaches, a compromise of these two represents another approach in which data sharing is coordinated by an open independent organizational body to provide recommendations and guidelines of a shared metadata and semantic structure, and the data management protocols to individual data owners prior to the sharing of the data. With this approach, sensitive data can be managed and de‐identified at the source, but at the same time, an agreeable set of higher‐level information of the individual subject‐level data can be pooled meaningfully at a cohort level. In addition, the coordinating organization can also suggest data access protocol to further accommodate access to the sensitive data for the right individuals who have the validated and verified permission to access those data. Global Alliance for Genomic Health24; an organization that enables genomic and clinical data sharing across federated networks, is one example of an organization that plays this role in recommending metadata standards, and the protocols for sharing federated sensitive data with different access permissions. Moreover, some research funders require that grantees submit a data management plan following specific data‐sharing principles (e.g., the European Research Council embraces the “The FAIR Guiding Principles for scientific data management and stewardship”).25
Figure 3.
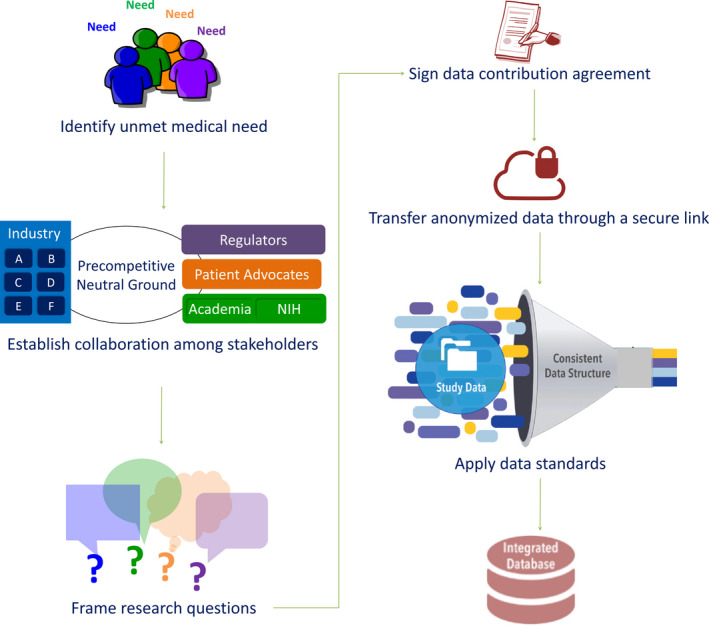
Stages for the creation of an integrated clinical database under an expanded data‐sharing approach (modified from22). NIH, National Institutes of Health.
At the Critical Path Institute (https://c‐path.org/), a nonprofit, public‐private partnership with the FDA, the contributor may choose different levels of disclosure: data can be made available to external researchers, consortium members only, or Critical Path Institute‐only. Once the data have been transferred, data mapping and integration takes place to yield a unified clinical trial database. A dedicated team remaps the nonstandardized data from individual studies by applying the standards from the Clinical Data Interchange Standards Consortium (CDISC) so that all data can be integrated into a single database. The CDISC (http://www.cdisc.org) is a nonprofit organization that develops data standards, methods, and tools for standardizing clinical trial data via common data elements as inputs into a database. CDISC Foundational Standards include Study Data Tabulation Model for clinical trial tables, Analysis Data Model for clinical trial analysis files, and others. An additional example of successful data sharing via technology‐enabled federated approaches (i.e., bringing analysis to the data) has been exercised successfully in the scope of the Beacon network within the Framework for Responsible Sharing of Genomic and Health‐related Data.26
The fast‐growing number of data‐sharing initiatives requires platforms that can map different initiatives worldwide. Consortia‐pedia (http://consortiapedia.fastercures.org) is a searchable catalog that includes a qualitative analysis of nearly 500 research consortia. The Global Alzheimer’s Association Interactive Network (http://www.gaain.org) is a platform that connects 51 Alzheimer’s disease (AD)‐related data repositories corresponding to over 400,000 subjects. Global Alzheimer’s Association Interactive Network’s Interrogator tool permits researchers to query and visualize data from different repositories. Such tools could facilitate collaboration and prevent duplication of efforts across consortia, besides enabling the use of clinical data to advance drug development, as explained in the following section.
Leveraging Individual Clinical Data
The leverage of publicly available clinical trials data has been successful in improving drug development by advancing drug development tools and allowing data repurposing to facilitate novel discoveries, as explained below.
Drug development tools
Drug development tools (DDTs) can be defined as methods, materials, or measures that have the potential to facilitate drug development and regulatory review process (e.g., biomarkers and clinical trial simulators). The FDA believes that the resources needed to develop a DDT for use across drug development programs are often beyond the capabilities of a single institution, encouraging collaboration among stakeholders (e.g., public‐private partnership). To support the advancement of DDT, the FDA established regulatory review and endorsement pathways for those27; for instance, (i) the Biomarker Qualification Program, with the goal of qualifying biomarkers for a specific context of use,28 and (ii) the Fit‐for‐Purpose Initiative, with the goal of endorsing dynamic tools that evolve over time upon availability of new data and methods (e.g., disease progression models and clinical trial simulators).29 Endorsed DDT up‐to‐date have typically resulted from collaboration, data, and knowledge sharing across stakeholders (Table 2).
Table 2.
List of drug development tools endorsed by the FDA with examples of data/knowledge sharing initiatives
| (Disease) Area | Endorsement | Data resource | Supporting information |
|---|---|---|---|
| Biomarker qualification | |||
| Nonclinical | Urinary biomarkers: Albumin, β2‐Microglobulin, Clusterin, Cystatin C, KIM‐1, Total Protein, and Trefoil factor‐3 | Short‐term rat GLP toxicology studies by Merck and Novartis | Guidance document: https://www.fda.gov/media/82532/download |
| Nephrotoxicity | Urinary nephrotoxicity biomarkers as assessed by immunoassays | Short term rat GLP toxicology studies conducted at AstraZeneca, Bayer, Biotrin, BMS, GSK, and Sanofi‐Aventis |
FDA review document: |
| Cardiac troponins | Serum/plasma cardiotoxicity biomarkers as assessed by immunoassay | Data from 20 publications as critical to the qualification of troponins for use in the rat |
FDA review document: |
| IS | Diagnostic biomarkers used with other clinical and host factors to identify patients with IS | The data to support qualification were obtained with the use of the Bio‐Rad Platelia Aspergillus enzyme immunoassay | FDA Guidance Document: https://www.fda.gov/media/94480/download |
| COPD | Prognostic biomarker used with other characteristics to enrich for COPD exacerbations | The COPD Biomarkers Qualification Consortium Database |
FDA Guidance Document: |
| PKD | Total kidney volume as assessed by magnetic resonance imaging, computed tomography, and ultrasound | Three patient registries (University of Colorado‐Denver, Mayo Clinic, and Emory University) and two longitudinal cohort studies (CRISP1 and CRISP2 on the natural history of autosomal dominant PKD | FDA Guidance Document: https://www.fda.gov/media/93105/download |
| Nephrotoxicity | Urinary nephrotoxicity biomarker panel as assessed by immunoassays | Normal healthy volunteers | Qualification Determination Letter: https://www.fda.gov/media/115635/download |
| CHMI | Monitoring biomarker informs initiation of treatment with antimalarial drug following CHMI with P. falciparum sporozoites in healthy subjects in clinical studies for vaccine and/or drug development | Nonclinical and clinical data from multiple sources including the University of Washington |
Qualification Determination Letter: |
| FFP initiative | |||
| AD | FFP Disease Progression Model: Placebo/Disease Progression | ADNI, and CPAD databases | Determination letter: https://www.fda.gov/media/98856/download |
| Multiple | FFP Statistical Method: MCP‐Mod | – | Determination letter: https://www.fda.gov/media/99296/download |
AD, Alzheimer’s disease; ADNI, AD Neuroimaging Initiative; CHMI, Controlled human malaria infection; COPD, chronic obstructive pulmonary disease; CPAD, Critical Path for Alzheimer's Disease; CRISP, Consortium for Radiologic Imaging Studies of Polycystic Kidney Disease; FDA, US Food and Drug Administration; FFP, Fit‐for‐Purpose; GLP, Good Laboratory Practice; IS, invasive aspergillosis; MCP‐Mod, Multiple Comparison Procedure – Modeling; PKD, Polycystic Kidney Disease.
The Critical Path for Alzheimer’s Disease Consortium ((CPAD), previously called Coalition Against Major Diseases, https://c‐path.org/programs/cpad/) developed a clinical trial simulation tool for mild‐to‐moderate AD.30, 31 The tool was endorsed by the FDA and the European Medicines Agency (EMA) through formal regulatory pathways (Fit‐for‐Purpose with the FDA and Qualification of Novel Methodologies with the EMA). To develop the tool, a model‐based meta‐analysis was conducted combining individual‐level data from the CPAD consortium (3,223 patients from clinical trial placebo arms) and ADNI (186 patients in an observational study32) databases, and summary‐level data from 73 literature references (representing 17,235 patients).30 The clinical trial simulation tool can simulate beyond the standard parallel design used in most phase II and III AD clinical trials and is still relevant, given the urgent need to better treat the millions of patients with mild‐to‐moderate AD.
Another effort from CPAD integrated longitudinal patient‐level data from two open‐access natural history studies in patients diagnosed with amnestic mild cognitive impairment (MCI)—ADNI‐1 and ADNI‐232to develop a neuroimaging‐informed clinical trial enrichment tool for amnestic MCI clinical trials. The clinical trial enrichment tool was defined as “a computer simulator that uses a disease progression model as a backbone – i.e., integrated information from the natural progression of the disease and individual patient characteristics that may be associated with differences in progression rate.”33 The tool showed that the inclusion of amnestic MCI subjects with baseline hippocampal volume less than the 84th or 50th percentile allowed an approximate reduction in trial size of at least 26% and 55%, respectively. This effort followed a similar approach to the one used before in the context of early‐stage Parkinson’s disease, demonstrating the utility of dopamine transporter neuroimaging as enrichment biomarker for clinical trials.34, 35, 36 Such effort was led by the Critical Path for Parkinson’s consortium, and used individual‐level longitudinal data of 672 subjects with early‐stage Parkinson’s disease in the disease in the Parkinson's Progression Markers Initiative observational study and the Parkinson Research Examination of CEP‐1347 Trial (PRECEPT) clinical trial. The analysis served as the basis for the EMA qualification of the dopamine transporter as an enrichment biomarker in that patient population.37 It constitutes a “model‐informed biomarker qualification,” to our knowledge, presented for the first time in the literature.34
Clinical trial data repurposing
With increased transparency in sharing the clinical research data, we are just beginning to explore the benefits of repurposing public data sets and generate data‐driven hypotheses that were not initially proposed in the studies. For instance, data repurposing initiatives from ImmPort (www.immport.org), a data warehouse that includes clinical trial subject‐level data from National Institute of Allergy and Infectious Diseases‐funded immunology studies, facilitated the translation of new insights into discoveries.38, 39
Using ImmPort public data, Nasrallah et al.40 sought to identify biomarkers for patients with antineutrophil cytoplasmic antibody‐associated vasculitis who failed treatment with cyclophosphamide or rituximab therapy. Their secondary analysis of rituximab‐treated participants in the Anti‐Neutrophil Cytoplasmic Antibodies‐Associated Vasculitis RAVE trial revealed distinct subsets of granulocytes at baseline in patients with vasculitis who achieved a complete remission with cyclophosphamide or rituximab treatment compared with patients who failed to achieve remission. Hence, this study suggested that profiling patients based on cell‐based markers might help to make therapeutic decisions and inform future trial designs.40
Another example of data repurposing includes the assessment of the post‐donation conditions for living donors in solid organ transplantation included in the ImmPort database. A curated data set, immTransplant, with post‐donation outcomes for kidney living donors were derived from 20 clinical studies (clinical trials and observational studies) in ImmPort.41 The availability of such data gives an opportunity to build a “trajectory map” for the sequence of events in a subset of living donors with a long‐term follow‐up post‐donation. This approach retrospectively helps to investigate the patterns of surgical and nonsurgical conditions that may arise post‐donation in living donors.
Despite these great examples and the well‐recognized benefits to be gained by sharing clinical trial data, there are several challenges and ethical considerations related to the sharing of clinical trial data that need to be addressed to make sure that all stakeholders involved receive maximum benefits while minimizing risks. These challenges will be discussed in the following sections, along with potential solutions.
Barriers Affecting Clinical Data Sharing
Several different barriers are affecting clinical trials data sharing. Some of those barriers are related to concerns of academic researchers who own the data, some are related to organizational policies implemented by research institutions that restrict data sharing, and others are related to technical challenges, privacy concerns, and ethical considerations that impede clinical trial data sharing.
Concerns about losing credit
For research scientists—especially early‐career researchers—peer‐review publication is the currency for academic advancement. Thus, a lot of researchers are not in favor of sharing data in the pursuit of maximizing the number of publications through subsequent analysis of their data set. To overcome this challenge, incentivizing researchers by acknowledging their scientific contribution and counting their data‐sharing efforts as criteria for their career advancement could motivate them to share their clinical research data.42 Receiving credit in the form of future grant support and formal recognition by research institutions can encourage academic researchers to share their clinical data and collaborate to facilitate data sharing. Additional solutions have also been proposed to provide appropriate recognition and credit for researchers who share their clinical data.43, 44, 45, 46 For instance, researchers who generated and shared their data could be designated as authors—data authors—on the manuscripts published via the reuse of their shared data.43 Additionally, several data repositories that generate a data citation, have been developed (i.e., Dryad and Harvard Dataverse).43 Proper citation of data and tracking data identifiers to know how many times a unique identifier of a data set has been shared via direct reuse or mapping/linking to other resources can help tracking the reuse of these data and provide recognition for creators, publishers, and distributors of these data. Moreover, many journals have now policies encouraging or mandating authors to provide data availability statements as well as citing the data sets used to give scholarly credit and legal attribution to all researchers who have contributed to generation and publishing of the data.47 All these efforts serve as solutions to give proper credit to clinical data holders and encourage them to collaborate and share their data to improve reproducibility and robustness of research findings and enable the reusability of data to address unmet clinical needs.
Privacy concerns and ethical considerations
The ethical arguments in favor of data sharing are overwhelming in two different, but inter‐related ways: first, the sheer volume of peer‐reviewed publications in favor of data sharing make it such that it is not a niche position of some bioethicists or group of scholars. For example, doing a PubMed search with the terms ‘“data sharing” AND “ethics” reveals 444 entries, most of which describe successful examples of data sharing, argue in favor of data sharing, or illustrate the difficulties related to data sharing and how best to overcome them. Second, and most importantly from an ethics viewpoint, the arguments that people use to defend and uphold the importance of data sharing are morally overdetermined (i.e., regardless of what ethical theory one espouses, everyone reaches the same conclusion in favor of data sharing). Thus, the first thing one must conclude is that from an ethics viewpoint, the need or duty to share data in clinical trials is unequivocally supported.
Most of the ethical issues that remain regarding clinical data sharing center around how to share data in a manner that is ethical without imposing patients involved in those trials to the risk of losing their privacy. To overcome this challenge, we need a clear, transparent, and accountable process that allows other researchers to use clinical trial data without affecting trial participants’ confidentiality and privacy. Developing extensive authentication and authorization infrastructure must be considered to provide strong privacy protection for trial participants. Analyzing the risks and benefits associated with data sharing over time and exploring newly emerging legal and technical tools to evaluate and mitigate risk of sharing clinical data overtime is needed. Adopting new technological solutions and proper Identity and Access Management systems that include privacy and security controls, such as de‐identification, ethical review processes, and secure data repositories are also needed to ensure stronger privacy protection to patient data.
Technical challenges
Secure data repositories that can store data and facilitate data queries are reasonable elements that need to be considered for building the required framework to facilitate responsible data sharing. These elements along with the recurring costs associated with data curation and data repositories, which need a reliable funding stream, are challenges that need to be addressed to assist in implementing a technological infrastructure that supports data sharing. Additionally, applying machine‐learning algorithms for data analysis tasks require a sufficiently large amount of training data from a wide range of possible scenarios; for example, multiple clinical trials, registries, and observational studies. The curation of such analysis data set is not trivial and can take a lot of resources and time, especially when pursuing regulatory endorsement of tools based on these data sets. Organizations, such as the Critical Path Institute,48 curates and hosts databases from a variety of different sources that can benefit researchers (e.g., CODR; https://codr.c‐path.org/). Advanced methods in computer vision involving imaging data usually require scans from many patients, sometimes over one hundred thousand. Performing large‐scale imaging, such as magnetic resonance imaging and computed tomography, might not be feasible and cost‐prohibitive for a single research group. These advanced methods would require concerted efforts from multiple research groups to come together and share data. Data sharing becomes even more critical for building DDTs that have a wide context of use. To overcome some of these technical challenges, we need to generate data‐sharing policies that are coupled with greater support and education for researchers in order to have faster and easier routes for sharing data optimally. Agencies, such as National Institute of Health and National Science Foundation, should provide a training program for researchers on best practices in data sharing. Additionally, explicit funding for data generation, management, and sharing tasks should also be provided. For instance, as part of a grant, funding could be set aside for building and maintaining the infrastructure for researchers to manage and share data.
Full interoperability and accessibility of clinical trial data are substantial for clinical trial data sharing. However, technical challenges related to inconsistency in clinical trial data collection and nonstandardized clinical trial data documentation are major challenges that impede the process of clinical data sharing. Additionally, with nonstandardized data collection formats, important information related to clinical trial design, patients drop out, and other complexities in the data may be missed during secondary analysis, which may lead to erroneous interpretations of the data. Conclusions from such analysis may challenge the original clinical trial findings and become a risk to the primary executors of the trial as well as the patients. Perhaps, one of the potential solutions to overcome these challenges is to impose standards for clinical trial data sharing. We can learn from the efforts made to standardize electronic health records data, where mechanisms and principals for standardization have been established and widely accepted. For instance, the Health Level 7 International developed Fast Healthcare Interoperability Resource; a protocol for standardizing healthcare data to improve interoperability and electronic transfer of data across the healthcare ecosystem. The Fast Healthcare Interoperability Resource requires data to be accessible, interoperable, and reusable to facilitate proper data reuse. Having a similar protocol for standardizing clinical trial data sharing, that stipulates a detailed and complete data annotation, will help in circumventing many data‐related obstacles and will allow for more usable trial data sharing.
Besides the concerns mentioned above, we also acknowledge additional issues related to metadata validation, long‐term sustainability of data, and the presence of organizational policies restricting data sharing. These concerns would need researchers from academia, industry, and legal authorities to collaborate and create appropriate channels to work closely on behalf of patients. Such collaborations would help overcome those challenges by generating the proper policies that would protect the privacy of trial participants and facilitate clinical trial data sharing and its reuse to address unmet patient needs.
Conclusions
We are currently witnessing continuous efforts to increase the transparency and breaking down the barriers of sharing clinical trials data between academic researchers, pharmaceutical industry, and legal authorities. Additionally, a cultural transformation supporting data sharing has already begun as leaders in academia, industry, and regulatory agencies started to embrace the value of data sharing and the breakdown of data silos to unleash the power of open data in achieving fast and informed decisions. Moreover, clinical trial participants have become more vocal in the request for greater clinical trial transparency and sharing of their data among research organizations to open unprecedented avenues for therapeutic breakthroughs. With all these changes, and with the tremendous amounts of clinical trial data available today, we believe that opening this treasure trove will have a significant influence on advancing scientific drug discovery and development and improving patient care.
As clinical pharmacologists, we must pave the way for clinical data sharing within our institutions and research communities in order to expedite the development of treatments for unmet medical needs. Possible ways of doing so include: (i) work on behalf of and with patients or patient advocacy groups to incentivize data sharing; (ii) ensure the inclusion of comprehensive data‐sharing plans within clinical study protocols, along with their subsequent execution; (iii) identify whether our institutions possess relevant clinical data that can be used to build DDT, and, if so, advocate for their sharing; (iv) demonstrate how open data can be used to increase efficiency of drug development programs; and (v) join public‐private partnerships to help the community to use data that may be familiar to you in a proper context. Together we can shape the future direction of clinical data sharing.
Funding
S.B. has funding support from the National Institute of Allergy and Infectious Diseases (ImmPort contract HHSN316201200036W) and Bill & Melinda Gates Foundation (OPP1196575).
Conflict of Interest
M.H.S. is an employee of Pfizer Inc. J.B. is an employee of the Critical Path Institute. J.P. is an employee of the Critical Path Institute. K.R. is an employee of the Critical Path Institute. D.J.C. was an employee of the Critical Path Institute. All other authors declared no competing interests for this work.
Author Contributions
All authors wrote the manuscript, designed the research, and performed the research.
References
- 1. The data deluge. Nat. Cell Biol. 14, 775 (2012). [DOI] [PubMed] [Google Scholar]
- 2. Qian, T. , Zhu, S. & Hoshida, Y. Use of big data in drug development for precision medicine: an update. Expert Rev. Precis. Med. Drug Dev. 4, 189–200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drazen, J.M. Sharing individual patient data from clinical trials. N. Engl. J. Med. 372, 201–202 (2015). [DOI] [PubMed] [Google Scholar]
- 4. Mello, M.M. et al Preparing for responsible sharing of clinical trial data. N. Engl. J. Med. 369, 1651–1658 (2013). [DOI] [PubMed] [Google Scholar]
- 5. Koenig, F. et al Sharing clinical trial data on patient level: opportunities and challenges. Biom. J. Biom. Z. 57, 8–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ross, J.S. & Krumholz, H.M. Ushering in a new era of open science through data sharing: the wall must come down. JAMA 309, 1355–1356 (2013). [DOI] [PubMed] [Google Scholar]
- 7. Krumholz, H.M. et al Sea change in open science and data sharing: leadership by industry. Circ. Cardiovasc. Qual. Outcomes 7, 499–504 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eichler, H.‐G. , Abadie, E. , Breckenridge, A. , Leufkens, H. & Rasi, G. Open clinical trial data for all? A view from regulators. PLoS Med. 9, e1001202 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawahara, T. , Fukuda, M. , Oba, K. , Sakamoto, J. & Buyse, M. Meta‐analysis of randomized clinical trials in the era of individual patient data sharing. Int. J. Clin. Oncol. 23, 403–409 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan, A.‐W. et al Increasing value and reducing waste: addressing inaccessible research. Lancet Lond. Engl. 383, 257–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doshi, P. , Jefferson, T. & Mar, C.D. The imperative to share clinical study reports: recommendations from the Tamiflu experience. PLoS Med. 9, e1001201 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. ClinicalTrials.gov Background – ClinicalTrials.gov. <https://clinicaltrials.gov/ct2/about‐site/background>.
- 13. Food and Drug Administration . Amendments Act of 2007 (2007).
- 14. Zarin, D. , Tse, T. , Williams, R. , Califf, R. & Ide, N. The ClinicalTrials.gov results database–update and key issues. N. Engl. J. Med. 364, 852–860 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hopewell, S. , Loudon, K. , Clarke, M. , Oxman, A. & Dickersin, K. Publication bias in clinical trials due to statistical significance or direction of trial results. Cochrane Database Syst. Rev. 1, MR000006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DeAngelis, C. et al Clinical trial registration: a statement from the international committee of medical journal editors. N. Engl. J. Med. 351, 1250–1251 (2004). [DOI] [PubMed] [Google Scholar]
- 17. Laine, C. et al Clinical trial registration–looking back and moving ahead. N. Engl. J. Med. 356, 2734–2736 (2007). [DOI] [PubMed] [Google Scholar]
- 18. Taichman, D. et al Data Sharing statements for clinical trials—a requirement of the international committee of medical journal editors. N. Engl. J. Med. 376, 2277–2279 (2017). [DOI] [PubMed] [Google Scholar]
- 19. Hopewell, S. , Clarke, M. , Stewart, L. & Tierney, J. Time to publication for results of clinical trials. Cochrane Database Syst. Rev. 2, MR000011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mello, M. , Lieou, V. & Goodman, S. Clinical trial participants’ views of the risks and benefits of data sharing. N. Engl. J. Med. 378, 2202–2211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilhelm, E.E. , Oster, E. & Shoulson, I. Approaches and costs for sharing clinical research data. JAMA 311, 1201–1202 (2014). [DOI] [PubMed] [Google Scholar]
- 22. Conrado, D.J. , Karlsson, M.O. , Romero, K. , Sarr, C. & Wilkins, J.J. Open innovation: Towards sharing of data, models and workflows. Eur. J. Pharm. Sci. Off. J. Eur. Fed. Pharm. Sci. 109S, S65–S71 (2017). [DOI] [PubMed] [Google Scholar]
- 23. TransCelerate . Data De‐identification and Anonymization of Individual Patient Data in Clinical Studies – A Model Approach. <http://www.transceleratebiopharmainc.com/wp‐content/uploads/2015/04/CDT‐Data‐Anonymization‐Paper‐FINAL.pdf>.
- 24. Global Alliance for Genomics and Health. GENOMICS . A federated ecosystem for sharing genomic, clinical data. Science 352, 1278–1280 (2016). [DOI] [PubMed] [Google Scholar]
- 25. European Research Council . Open Research Data and Data Management Plans. <https://erc.europa.eu/sites/default/files/document/file/ERC_info_document‐Open_Research_Data_and_Data_Management_Plans.pdf>.
- 26. Fiume, M. et al Federated discovery and sharing of genomic data using Beacons. Nat. Biotechnol. 37, 220–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Center for Drug Evaluation and Research . Drug Development Tool Qualification Programs. FDA <http://www.fda.gov/drugs/development‐approval‐process‐drugs/drug‐development‐tool‐qualification‐programs> (2019).
- 28. Center for Drug Evaluation and Research . CDER Biomarker Qualification Program. FDA <http://www.fda.gov/drugs/drug‐development‐tool‐qualification‐programs/cder‐biomarker‐qualification‐program> (2019).
- 29. Center for Drug Evaluation and Research . Drug Development Tools: Fit‐for‐Purpose Initiative. FDA (2019).
- 30. Rogers, J.A. et al Combining patient‐level and summary‐level data for Alzheimer’s disease modeling and simulation: a β regression meta‐analysis. J. Pharmacokinet. Pharmacodyn. 39, 479–498 (2012). [DOI] [PubMed] [Google Scholar]
- 31. Romero, K. et al The future is now: model‐based clinical trial design for Alzheimer’s disease. Clin. Pharmacol. Ther. 97, 210–214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weiner, M.W. et al The Alzheimer’s disease neuroimaging initiative: a review of papers published since its inception. Alzheimers Dement. 8, S1–S68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Conrado, D.J. et al Hippocampal neuroimaging‐informed clinical trial enrichment tool for amnestic mild cognitive impairment using open data. Clin. Pharmacol. Ther. 107, 903–914 (2020). [DOI] [PubMed] [Google Scholar]
- 34. Conrado, D.J. et al Dopamine transporter neuroimaging as an enrichment biomarker in early Parkinson’s disease clinical trials: a disease progression modeling analysis. Clin. Transl. Sci. 11, 63–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romero, K. et al Molecular neuroimaging of the dopamine transporter as a patient enrichment biomarker for clinical trials for early Parkinson’s disease. Clin. Transl. Sci. 12, 240–246 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stephenson, D. et al The qualification of an enrichment biomarker for clinical trials targeting early stages of Parkinson’s disease. J. Park. Dis. 9, 553–563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qualification opinion on dopamine transporter imaging as an enrichment biomarker for Parkinson’s disease clinical trials in patients with early Parkinsonian symptoms. <https://www.ema.europa.eu/en/documents/regulatory‐procedural‐guideline/qualification‐opinion‐dopamine‐transporter‐imaging‐enrichment‐biomarker‐parkinsons‐disease‐clinical_en.pdf>.
- 38. Bhattacharya, S. et al ImmPort: disseminating data to the public for the future of immunology. Immunol. Res. 58, 234–239 (2014). [DOI] [PubMed] [Google Scholar]
- 39. Bhattacharya, S. et al ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 5, 180015 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nasrallah, M. et al Reanalysis of the Rituximab in ANCA‐Associated Vasculitis trial identifies granulocyte subsets as a novel early marker of successful treatment. Arthritis Res. Ther. 17, 262 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen, J. et al Assessment of postdonation outcomes in US living kidney donors using publicly available data sets. JAMA Netw. Open 2, e191851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moher, D. et al Assessing scientists for hiring, promotion, and tenure. PLoS Biol. 16, e2004089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bierer, B.E. , Crosas, M. & Pierce, H.H. Data authorship as an incentive to data sharing. N. Engl. J. Med. 376, 1684–1687 (2017). [DOI] [PubMed] [Google Scholar]
- 44. Gewin, V. Data sharing: an open mind on open data. Nature 529, 117–119 (2016). [DOI] [PubMed] [Google Scholar]
- 45. Popkin, G. Data sharing and how it can benefit your scientific career. Nature 569, 445–447 (2019). [DOI] [PubMed] [Google Scholar]
- 46. Stall, S. et al Make scientific data FAIR. Nature 570, 27–29 (2019). [DOI] [PubMed] [Google Scholar]
- 47. Cousijn, H. et al A data citation roadmap for scientific publishers. Sci. Data 5, 180259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. C‐Path . About | Critical Path Institute. <https://c‐path.org/about/>.
- 49. Burton, J. , Bhattacharya, S. , Romero, K. & Conrado, D.J. Open data for clinical pharmacology. Clin. Pharmacol. Ther. 107, 703–706 (2020). [DOI] [PubMed] [Google Scholar]
- 50. Imperial, M.Z. et al A patient‐level pooled analysis of treatment‐shortening regimens for drug‐susceptible pulmonary tuberculosis. Nat. Med. 24, 1708–1715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


