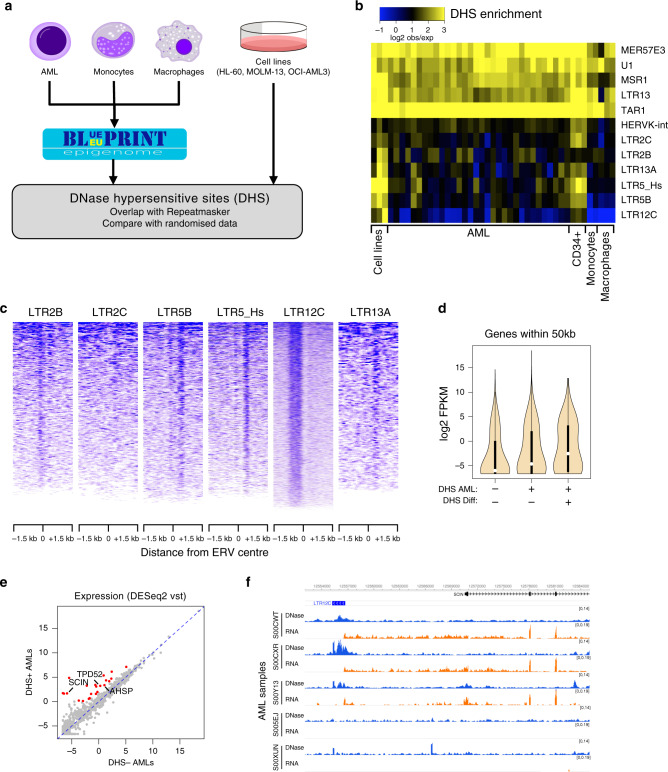
Fig. 1. ERVs with regulatory potential are activated in AML.
a Schematic of the strategy to detect repeat families associated with open chromatin in AML (hematopoietic cells’ credit: A. Rad and M. Häggström; CC-BY-SA-3.0 licence). b Heatmap of the observed/expected enrichment for DHSs in selected repeat families. Cell lines are presented in the following order: HL-60, MOLM-13 and OCI-AML3. c DNase-seq profile across all elements of each AML DHS-associated repeat (A-DAR) families in OCI-AML3. d Gene expression average across all Blueprint AML samples for genes within 50 kb of A-DARs with or without a DHS in AML and/or in differentiated cells (boxes indicate first, second (median) and third quartiles; whiskers indicate data within 1.5× of the interquartile range). e For each gene lying near an A-DAR element, we compared its expression in AML samples (n = 26) where the respective ERV has a DHS, versus AML samples where the DHS is absent. Expression values were normalised using the variance-stabilising transformation (vst, log2 scale) in DESeq2. Highlighted are genes with >4-fold difference and vst > 0. f Example of a gene (SCIN) that displays a strict correlation between its expression (orange) and the presence of a DHS peak (blue) at a nearby LTR12C element in different AML samples.