Figure 2.
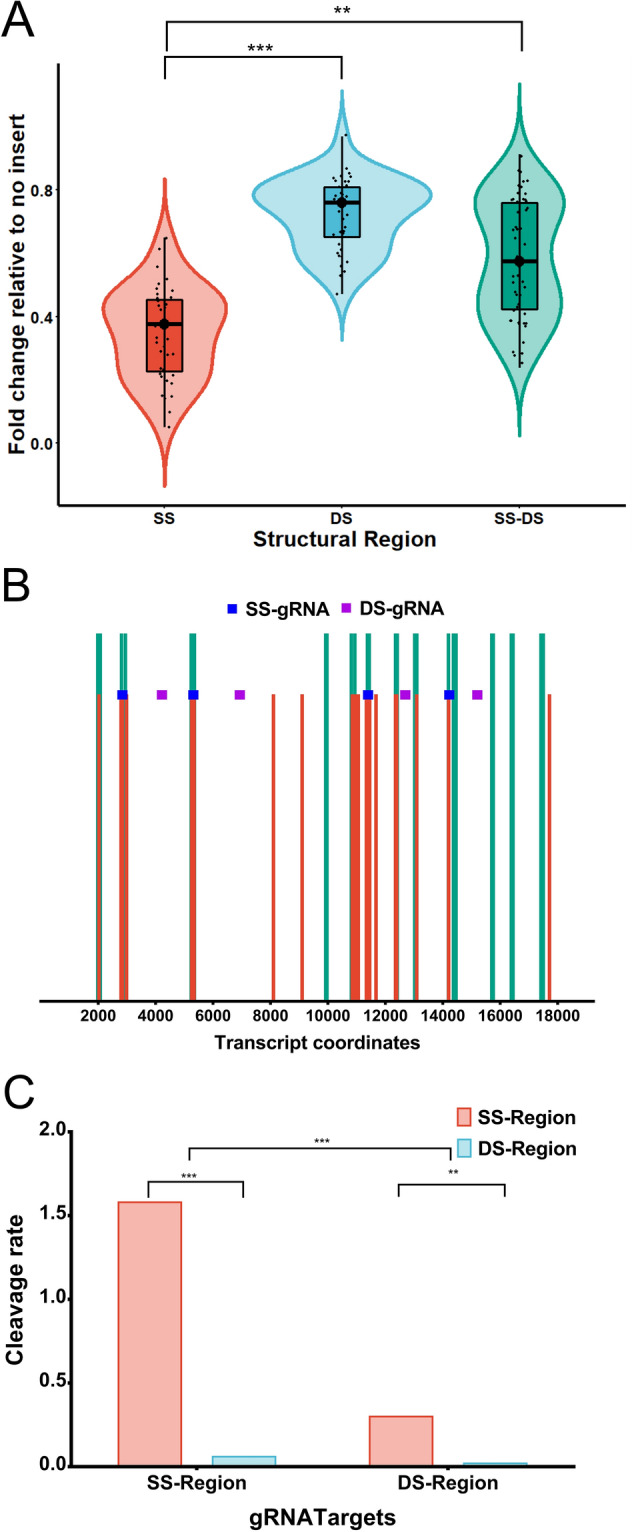
XIST knockdown upon gRNA targeting the SS and DS regions in the transcript. (A) Set of gRNAs targeting the SS regions (loops) brings about a significant knockdown of XIST transcript compared to the ones targeting the DS regions (stems). gRNAs targeting the SS-DS regions (stem-loop junctions), however, demonstrate wide expression range having few gRNAs efficient for transcript knockdown. (B) The red vertical bars indicate the cleaved locus in the transcript as analysed from RNA-seq and HISAT 2.0 mapping. Green vertical bars indicate SS regions; annotated in reference to the structure-seq data. A significant overlapping of the cleaved nucleotides onto the SS regions shows the selective cleavage preference of Cas13 endonucleases for SS regions. (SS-gRNA and DS-gRNA respectively target SS and DS regions) (C) Cleavage rate in an RNA-seq experiment was determined by the ratio of cleaved nucleotides in a given transcript region (SS or DS) to the uncleaved nucleotides. Note that the cleavage rate is significantly high for SS regions compared to DS regions. Although certain gRNAs directed for SS regions show a minor and unspecific cleavage at DS regions and vice versa, nevertheless their number was insignificant compared to gRNAs those demonstrating strand-specific cleavage.
