Figure 3.
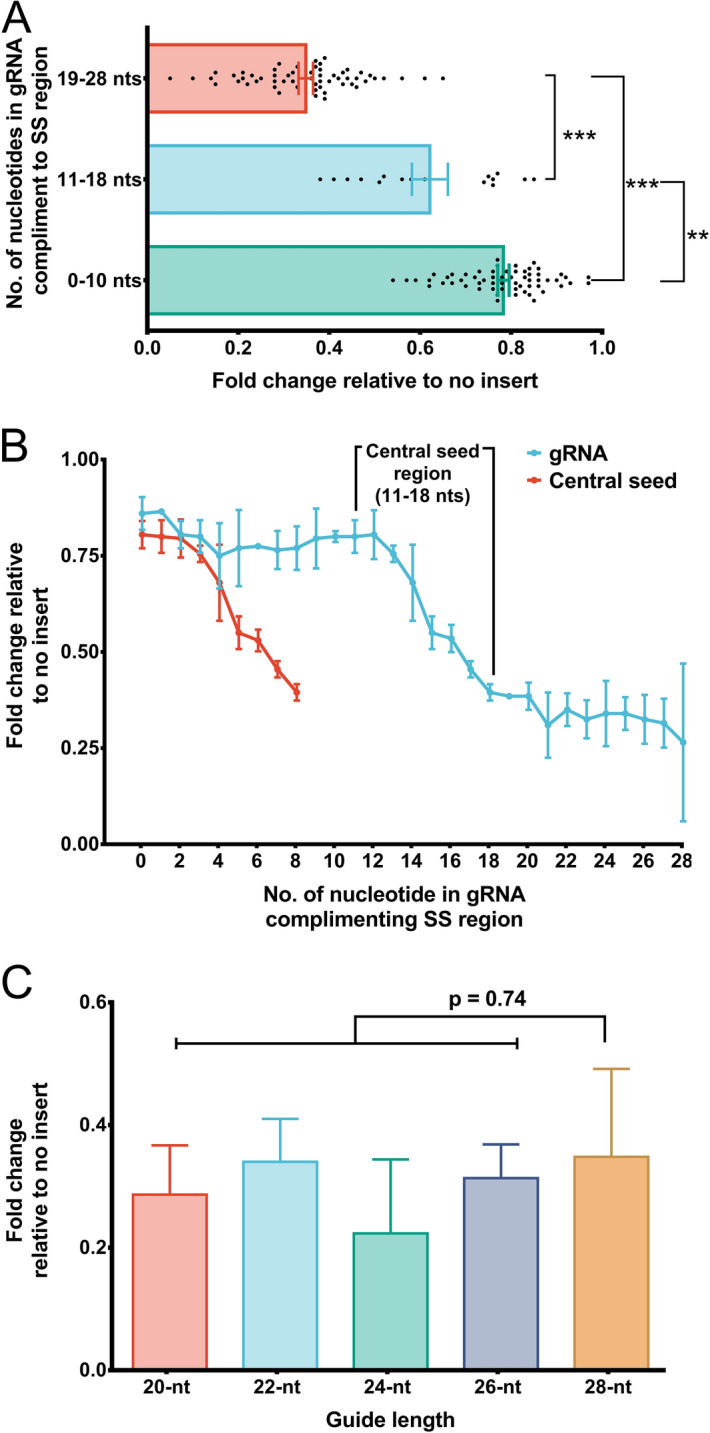
Requirement of single-stranded nucleotides in the transcript for Cas13 activity. (A) 28 nt gRNAs stratified into 3 subsets depending upon their complementarity to nucleotides in the SS region. Among the three subsets, gRNAs with 0–10 nts complementing nucleotides of SS regions bring about poor knockdown of the transcript which is evident from high expression values. For the subset of gRNAs that complement SS region nucleotides with 11–18 nts, a wide range of transcript expression values was observed. This subset comprised only a few gRNAs that induced efficient knockdown (gRNAs which complemented SS regions nucleotides with 17–18 nts). The subset of gRNAs complementing the SS regions with 19–28 nts was the most efficient among the three subsets to induce knockdown. (B) The extent of transcript knockdown analyzed in relation to the number of each nucleotide in guide RNA that complement the nucleotides in SS regions in SS-DS junctions. gRNAs with 0–10 nts complementing SS regions bring about poor knockdown, however, transcript expression declines with gRNAs those complementing nucleotides of SS regions starting with 11 nts reaching up to 18 nts. While, gRNAs with more than (and including) 18 nts (18–28 nts) complementing SS region nucleotides demonstrated superior knockdown of the transcript. It is, in fact the, the “central seed” binding region in the gRNA with 8 central bases (11–18 nts) which upon complementing SS region in the transcript brings about significant knockdown. Moreover, when complementarity exceeds the central seed bases, there was no apparent change in the transcript knockdown. This implies that, the central seed region in the gRNAs upon complementing SS regions is sufficient to promote Cas13 mediated transcript knockdown. (C) An insignificant change in the XIST expression upon targeting with 20–28 nts long gRNAs shows that Cas13 mediated transcript knockdown is tolerated by varying gRNA lengths, provided their central seed bases target the SS regions of the transcript.
