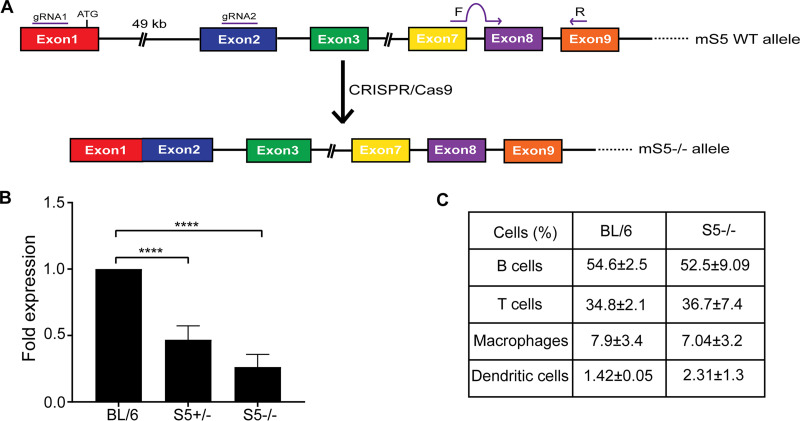
FIG 2.
SERINC5−/− mice have lower levels of mSERINC5 and retain normal populations of leukocytes. (A) Schematic of the SERINC5 allele in the SERINC5−/− mice. Two guide RNAs (gRNAs) were used to target exons 1 and 2. The genomic fragment between the two gRNAs was excised using CRISPR/Cas9 technology, resulting in the abrogation of the SERINC5 gene. Locations of the SERINC5 primers used for RT-PCR analysis are indicated by the “F” and “R” purple arrows. (B) Fold change of SERINC5 expression in the SERINC5−/− and SERINC5+/− mice relative to C57BL/6 mice. SERINC5 RNA levels in the SERINC5−/− mice (n = 5), SERINC5+/− mice (n = 6), and C57BL/6 mice (n = 6) were determined by RT-qPCR and normalized to β-actin. (C) PBMCs from 4 C57BL/6 mice and 6 SERINC5−/− mice were stained with anti-CD45R/B220 (B cells), anti-CD3 (T cells), anti-F4/80 (Macrophages), and anti-CD11c (Dendritic cells) antibodies and subjected to FACS analysis. Values are presented as means ± SD. There were no significant differences in the percentages of T cells, B cells, macrophages, and dendritic cells among the C57BL/6 and SERINC5−/− mice. For the data shown in panel B, statistical significance was determined by one-way ANOVA and Tukey’s test; for the data shown in panel C, statistical significance was determined by an unpaired t test, **, P < 0.01; ****, P < 0.0001. S5, SERINC5; BL/6, C57BL/6.