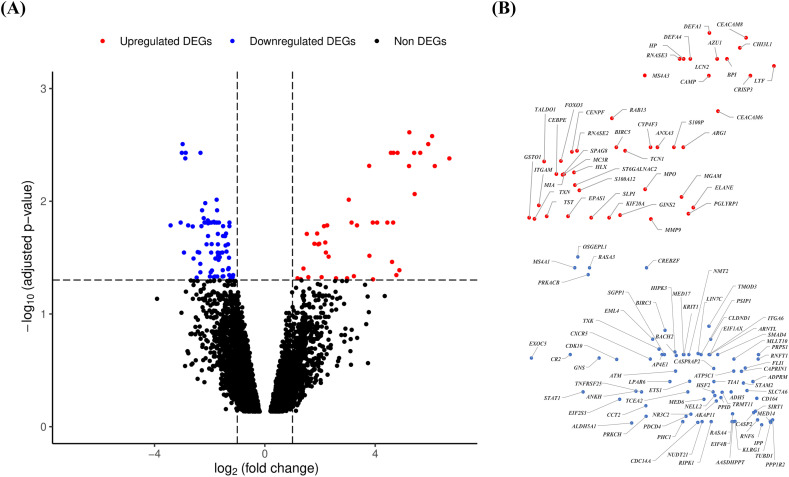
Fig. 2.
Volcano plot analysis to identify the differentially expressed genes (DEGs). The expressions of genes are evaluated by analysis of microarray data of the peripheral blood samples of SARS-CoV patients (n = 10) versus healthy controls (n = 4) collected from the data source (GSE1739) using Bayesian algorithm in limma Bioconductor package of bioinformatics tools in R language and environment. A: The volcano plot of the expressions of genes using the logarithmic values of fold changes (log2 fold change) in x-axis and ‘false discovery rate’ (FDR) adjusted p-values (−log10 adjusted p-value) in y-axis. Dotted lines parallel to x-axis and y-axis indicate the threshold values using FDR adjusted p-value < 0.05 and | log2 fold change | > 1 respectively to identify the upregulated and downregulated DEGs. The red, blue and black color dots indicate the upregulated DEGs, downregulated DEGs and non DEGs respectively. B: The gene IDs of upregulated (upper panel) and downregulated (lower panel) DEGs found in volcano plot. The position of dots of DEGs (45 upregulated and 75 downregulated) are same as those appeared in volcano plot. The scales of upregulated and downregulated DEGs are adjusted manually for proper presentation of the gene IDs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)