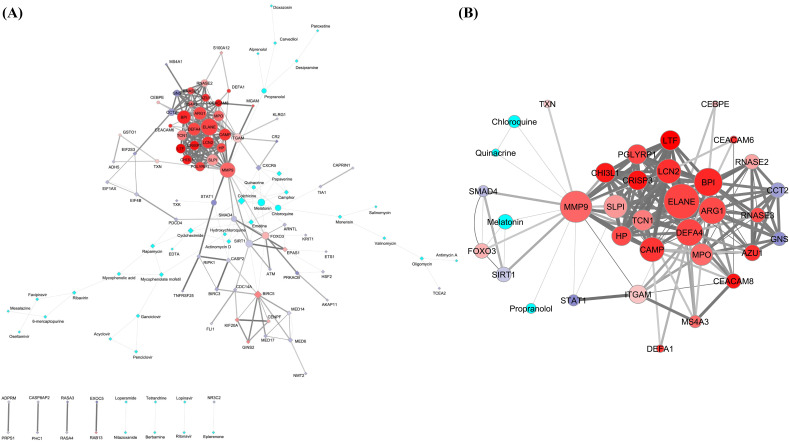
Fig. 3.
The interactome models of PPI-CPI network (A) and its top ranked sub-network (B) obtained in the Cytoscape software. The networks are developed using DEGs of the SARS patients and the promising COVID-19 repurposed drug candidates. A: The PPI-CPI network consists of 118 nodes of gene products/proteins and drugs corresponding to their respective gene IDs (35 upregulated and 43 downregulated DEGs) and name of drugs (40 in number). The 293 edges correspond to the functional connectivities between nodes. The upper panels of the images represent networks having continuous connections. The lower panels of the same designate discrete networks having only single connections. B: The top ranked sub-network was identified in the MCODE module of the Cytoscape considering MCODE score ≥ 4. The sub-network consists of 35 nodes of gene products/proteins and drugs of corresponding gene IDs (26 upregulated and 5 downregulated DEGs) and name of drugs (4 in number) respectively. The 174 edges correspond to the functional connectivities between nodes. A and B: The shape of the nodes in interactome models (A and B) depicted as circles represent the nodes associated with the top ranked sub-network. The rest of the nodes in PPI-CPI network (A) are manually altered to diamond shapes to identify the top ranked sub-network in the main network for better interpretation. The intrinsic attributes of nodes and edges are kept intact in interactome models (A and B). The border of the nodes is illustrated with 2 pts. grey color. The sizes of the nodes indicate their connectivities (higher the value, higher will be the size) adjusted by the ‘continuous mapping of node size’ in ranges between 25 and 60 pts. for the lowest and highest node size respectively. The color of the nodes is represented as cyan for the drugs. The color gradients of red:white:blue are denoted as the expression pattern (log2 fold change) of genes specific for gene products/proteins (nodes), whereby the gradients of the colors have been adjusted to the expression values over the ranges of 6.68:0:−6.68 from the ‘continuous mapping of node color’ in the node network style of the Cytoscape. The widths of the edges are based on the combined scores (0.600 to 1) obtained as interaction data in the STRING for PPI network and STITCH for CPI network which are adjusted by 0.5 to 5 pts. of ‘continuous mapping of edge width’ in the edge network style of the Cytoscape. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)