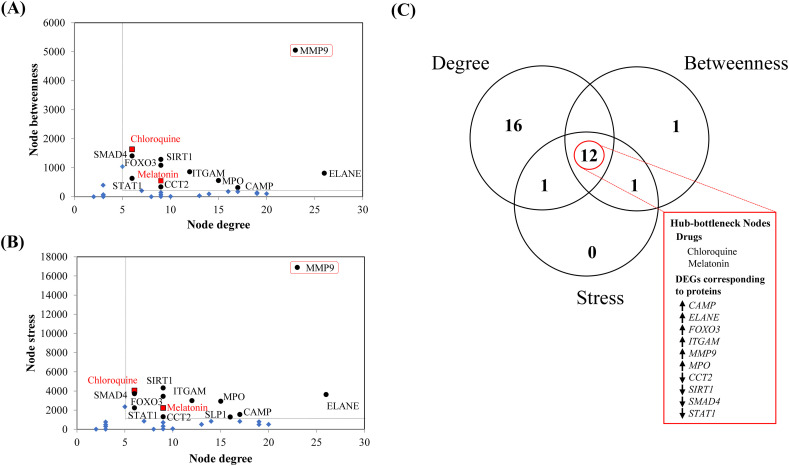
Fig. 4.
Pictorial summary of the topological properties and the centrality analyses for the top ranked sub-network to identify the hub-bottleneck nodes (A, B, C) using the CentiScaPe module of Cytoscape software. The identification of nodes of gene products/proteins and drugs are designated by corresponding gene IDs and name of the drugs. Graphical plots represent the dot plots of values of (A) node degree (x-axis) vs. node betweenness (y-axis), (B) node degree (x-axis) vs. node stress (y-axis) and (C) Venn diagram of high node degree/connectivity, high node betweenness and high node stress. Here, the term ‘high’ indicates higher than the mean cut-off thresholds for node degree/connectivity, betweenness and stress, which have been obtained from the CentiScaPe module of the Cytoscape software. Mean centrality values are presented as dotted lines in the graphs (A, B). The black round dots are hub-bottleneck protein nodes, the blue diamond shapes are non-hub-bottleneck nodes (proteins and drugs) and red boxes with black borders are the hub-bottleneck drug nodes in the (A, B) graphs. MMP9 in the red outlined boxes in the graph (A, B) represent the only target of chloroquine and melatonin among other hub-bottleneck nodes of the top-ranked sub-network. (C) Venn diagram indicates the common nodes that have the topological centrality indices viz. node degree/connectivity, betweenness and stress with the values higher than the mean cut-off respective threshold values obtained from the CentiScaPe module of the Cytoscape software. The upward and downward arrows indicate the expressions of upregulated and downregulated genes (gene IDs right to the arrows) corresponding to respective gene products/proteins in the hub-bottleneck nodes of COVID-19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)