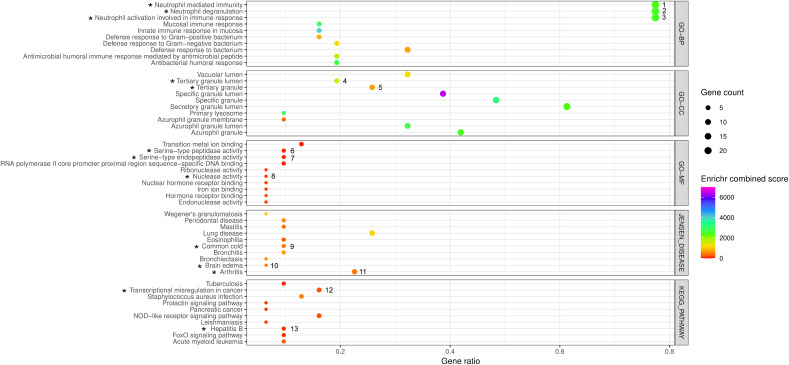
Fig. 5.
The bubble plot to demonstrate the summary of the functional enrichment analysis of DEGs under the top ranked sub-network cluster of the PPI-CPI network in COVID-19 identified by the Enrichr program. The results include the top ten enriched terms corresponding to each of the Enrichr functional annotations viz. GO terms [Biological Process (GO-BP), Cellular Component (GO-CC), Molecular Function(GO-MF)], KEGG pathways and Jensen Diseases obtained from Enrichr analyses using 31 genes corresponding to the gene products/proteins nodes of top ranked sub-network of the PPI-CPI network in COVID-19. The abscissa represents the ‘Enriched gene ratio’. The sizes of bubbles indicate the ‘Enriched gene count’ adjusted with 0.5 to 4 pts. based upon the gene counts of respective functional annotations. The colors of the bubbles indicate the ‘Enrichr combined scores’ (log(p-value) × z-value) which are adjusted in a color (VIBGYOR) gradient, ranging from 0 to 7000 on the basis of values of the scores. The p-value < 0.05 of Fisher's exact test is considered for significant result. The black colored asterisks indicate the enriched functional annotations of the gene products/proteins including MMP9 that correspond to their DEGs denoted by numbers (1 to 13). 1–3: ARG1, AZU1, BPI, CAMP, CCT2, CEACAM6, CEACAM8, CHI3L1, CRISP3, DEFA1, DEFA4, ELANE, GNS, ITGAM, LCN2, LTF, MMP9, MPO, MS4A3, PGLYRP1, RNASE2, RNASE3, SLPI, TCN1; 4: CAMP, CRISP3, LTF, MMP9, PGLYRP1, TCN1; 5: CAMP, CEACAM8, CRISP3, ITGAM, LTF, MMP9, PGLYRP1, TCN1; 6–7: AZU1, ELANE, MMP9; 8: RNASE2, RNASE3; 9: MMP9, MPO, RNASE3; 10: MMP9, MPO; 11: ITGAM, STAT1, HP, CHI3L1, MPO, ELANE, MMP9; 12: CEBPE, ELANE, ITGAM, MMP9, MPO; 13: MMP9, SMAD4, STAT1.