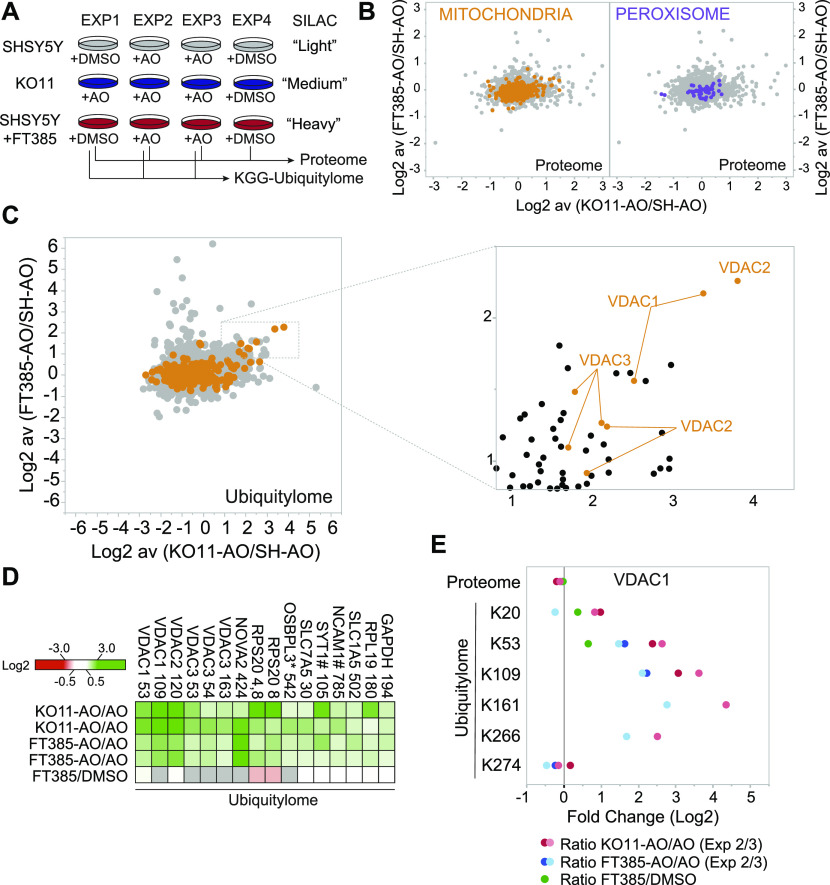
Figure 4. Comparison of proteome and ubiquitylome changes in USP30 KO versus USP30 inhibitor treated SHSY5Y cells.
(A) Schematic flow chart of SILAC based quantitative ubiquitylome and proteome analysis comparing USP30 KO and USP30 inhibition. SHSY5Y (USP30 wild-type) and SHSHSY USP30 KO (KO11) cells were metabolically labelled by SILAC as shown. Cells were then treated for 24 h with DMSO or antimycin A and oligomycin A (AO; 1 μM each) and/or FT385 (200 nM) as indicated. Cells were lysed and processed for mass spectrometry analysis. (B, C) Graphs depicting the fold change (log2) in the proteome (B) or ubiquitylome (C) of AO-treated SHSY5Y cells ± FT385 treatment (y-axis) and ± USP30 (x-axis). Mitochondrial (Integrated Mitochondrial Protein Index database; http://www.mrc-mbu.cam.ac.uk/impi; “known mitochondrial” only) and peroxisomal proteins (peroxisomeDB; http://www.peroxisomedb.org) proteins are highlighted in orange and purple, respectively. Inset in (C) shows enlarged section of ubiquitylome data for peptides enriched in USP30 KO and inhibitor treated cells. (B) Within proteome graphs (B) each dot represents a single protein identified by at least two peptides and the ratio shows the average of two experiments. (C) Within ubiquitylome graphs (C) each dot represents a single diGly peptide (localisation ≥ 0.75) and the ratio shows the average of two experiments. (D) Heat map showing diGly peptides that are increased consistently by log2 ≥ 0.8 in both USP30 KO and USP30 inhibitor (FT385) treated cells. Grey indicates the protein was not seen in that condition, * indicates ambiguity of peptide assignment between family members (OSBPL3, OSBPL7, and OSBPL6), # indicates an increase at proteome level in KO11. VDAC3 K53 and K54 correspond to equivalent lysines in two distinct isoforms. (E) Fold change (log2) in proteome and individual diGly peptides (localisation ≥ 0.75) by site in VDAC1 proteins. (D) See Fig S3 for corresponding data sets for VDAC2 and 3 and proteome data for hits shown in (D).