Figure 1.
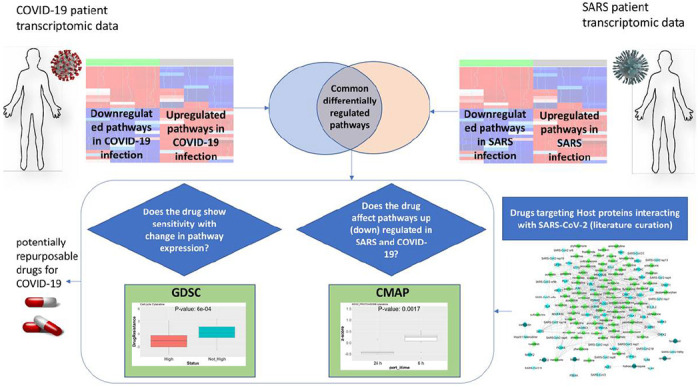
Graphical description of our approach to systematically assess candidate drugs that can be re-purposed for Covid-19 (or SARS) treatment, based on the transcriptomic changes of infected patients. We combined recently published COVID-19-infected patient transcriptomic data and previous SARS-infected patient transcriptomic data, to find key pathway alterations in infected patients. On the other hand, we collected the drugs targeting these SARS-CoV-2/SARS-CoV-1-interacting proteins from DrugBank and DGIdb to assess their effect in COVID-19- and SARS-infected patients, in-silico. Next, using the available drug perturbational data sets from the Broad Institute CMAP project, we assessed how the candidate drugs targeting SARS-CoV-2-interacting proteins affect the pathways that are altered after COVID-19 or SARS infection in a time-dependent manner. Using another available pharmacogenomic data resource, Genomics of Drug Sensitivity in Cancer (GDSC), we assessed whether the altered pathways affect sensitivity or resistance to our candidate drugs in human cell lines. Finally, we present a list of potential drugs for COVID-19 treatment that affect the COVID-19-altered pathways and incur less resistance in the presence of alteration in the pathways.
