Figure 5.
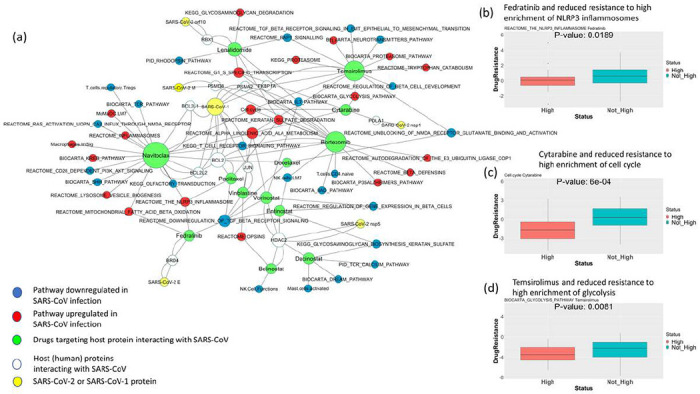
(a) The network shows the repurposable drugs associated with reduced resistance to the pathways upregulated or downregulated in SARS and COVID-19 infection. Interactions of SARS-Cov-1 or SARS-Cov-2 with host proteins are collected from Gordon et al.; the drugs targeting the host proteins are collected from DrugBank and DGIdb10,11; and the pathways upregulated or downregulated in COVID-19 infection that makes cell lines less resistant to the drugs targeting SARS-Cov-2-interacting proteins are calculated using cell line drug screening and cell line transcriptomic data from GDSC. (b-d) The boxplots show the differences in resistance (IC50) to the drugs (b) fedratinib, (c) cytarabine, and (d) temsirolimus in cell lines of hematopoietic or lymphoid origin from GDSC when (b) NLRP3 inflammosomes, (c) cell cycle, and (d) glycolysis are highly enriched (scaled ssGSEA score > 2) vs. not enriched. For pathways downregulated in COVID-19-infected patients, the cell line drug resistance is compared between cells where the pathway has low enrichment (scaled ssGSEA < −2) vs. other cells.
