Abstract
Hyperglycaemia can strongly alter the epigenetic signatures in many types of human vascular cells providing persistent perturbations of protein–protein interactions both in micro- and macro-domains. The establishment of these epigenetic changes may precede cardiovascular (CV) complications and help us to predict vascular lesions in diabetic patients. Importantly, these epigenetic marks may be transmitted across several generations (transgenerational effect) and increase the individual risk of disease. Aberrant DNA methylation and imbalance of histone modifications, mainly acetylation and methylation of H3, represent key determinants of vascular lesions and, thus, putative useful biomarkers for prevention and diagnosis of CV risk in diabetics. Moreover, a differential expression of some micro-RNAs (miRNAs), mainly miR-126, may be a useful prognostic biomarker for atherosclerosis development in asymptomatic subjects. Recently, also environmental-induced chemical perturbations in mRNA (epitranscriptome), mainly the N6-methyladenosine, have been associated with obesity and diabetes. Importantly, reversal of epigenetic changes by modulation of lifestyle and use of metformin, statins, fenofibrate, and apabetalone may offer useful therapeutic options to prevent or delay CV events in diabetics increasing the opportunity for personalized therapy. Network medicine is a promising molecular-bioinformatic approach to identify the signalling pathways underlying the pathogenesis of CV lesions in diabetic patients. Moreover, machine learning tools combined with tomography are advancing the individualized assessment of CV risk in these patients. We remark the need for combining epigenetics and advanced bioinformatic platforms to improve the prediction of vascular lesions in diabetics increasing the opportunity for CV precision medicine.
Keywords: Diabetic vasculature, Epigenetics, Cardiovascular prevention, Network medicine, Machine learning, Personalized therapy
Introduction
Diabetes refers to a group of metabolic chronic diseases sharing some hyperglycaemia-induced intermediate phenotypes, including systemic inflammation, oxidative stress, hypercoagulability, and endothelial dysfunction, which play an important role in the development of vascular complications in diabetic patients.1,2 Remarkably, vascular lesions in both micro- and macro-domains may be prevented as well as delayed in their progression when detected at early stages in asymptomatic subjects.1,2 This objective may be achieved by identifying novel biomarkers predicting and monitoring the progression of diabetes and its long-term complications. DNA polymorphisms can modulate the individual cardiovascular (CV) risk as well as the response to drug therapy. Some useful genome-wide polygenic score platforms are now available3,4; however, pharmacogenomics is still not a reality in the clinical market. Whereas genetics alone can explain a part of the inherited pathogenic mechanisms, changes in the epigenome may illuminate a significant fraction of the ‘missing hereditability’ by clarifying the complex relationship between genome and environment in modulating the risk and progression of lesions in diabetic vasculature.5 Transient hyperglycaemia can trigger an early chromatin remodelling in different types of vascular cells, which persists even when patients return to euglycaemic state inducing a maladaptive response, known as ‘metabolic memory’.6 Epigenetic regulatory mechanisms include DNA and RNA methylation, histone and non-histone post-translational modifications, and noncoding RNAs.7,8 Unlike DNA variations, aberrant epigenetic changes are pharmacologically reversible after treatment with specific small agents, known as ‘epidrugs’ (e.g. metformin and statins) which might prevent or revert the persistent effect of metabolic memory.7,8 Mapping the epigenomic fingerprint of the diabetic vasculature may be really advantageous to provide novel useful predicting biomarkers and drug targets, in order to delineate a personalized picture of the CV risk including prevention and/or rehabilitation programmes.8 This review updates on the epigenetic-sensitive mechanisms predicting the CV risk in diabetic patients, with a particular focus on the main epidrugs in clinical trials. Furthermore, we offer a clinical perspective where epigenetics integrated with imaging tools and network medicine could help to refine the assessment of CV risk leading to personalized therapy of diabetic patients.5,9
A focus on epigenetic signatures
DNA methylation
DNA methylation is catalysed by three groups of DNA methyltransferases (DNMTs): DNMT1, 2, or 3, which act at the carbon-5′ position of cytosines into ‘CpG islands’ by using S-adenosylmethionine as donor of the methyl group.9 DNA methylation mainly acts at transcriptional level by reducing the accessibility of transcription machinery to gene promoters.9 A class of demethylases belonging to 10–11 translocation (TET) family of DNA dioxygenases (TET1/2/3) controls the genome methylation pattern by oxidizing 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC).9 Depending on a biochemical cycle that requires methyl donors, DNA methylation can be reverted by using targeted dietary interventions (e.g. folic acid and vitamin B6, B12) or treatment with DNA methylation inhibitors, such as 5-aza-2-deoxycytidine. Thus, aberrant DNA methylation may be a useful drug target to prevent or modulate the progression of diabetes.10
Post-translational modifications
Post-translational modifications include histone and non-histone protein changes. Acetylation at specific amino-acid positions is the most studied in diabetic vasculature.6,10 The global acetylation level is regulated by two families of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDACs), which add and remove acetyl groups mainly at lysine residues of histone tails with activating and repressive effects, respectively.11 Some classes of HDAC inhibitors, such as trichostatin A, as well as HDAC activators, such as metformin, show anti-inflammatory properties that may provide useful additional drug strategies in diabetes management.10
MicroRNA
MicroRNA (miRNAs) are a family of small non-coding RNA molecules (21–22 nucleotides) regulating gene expression by binding to specific mRNAs. As downstream effect, miRNAs block their translation or induce their degradation.12 MiRNAs are originally transcribed from precursors in the nucleus to reach their final mature structure into the cytoplasm where they are assembled into miRNA-induced silencing complexes (miRSCs), which regulate the expression of several genes influencing the function of the parental cell.12 Moreover, they can also be secreted into microvesicles or exosomes, acting as signalling molecules and participating in intercellular cross-talk communication.12 MiRNAs are remarkably stable and their detection is repeatable and non-invasive, representing useful and early biomarkers for disease diagnosis, prevention, and prognosis.13 Several limitations due to isolation techniques, types of sample and study methodology are still ongoing and challenge the translation of basic findings in clinical arena. Furthermore, miRNA-based therapeutic strategy may have a great clinical potential, owing to the ability to deliver oligonucleotides to mimic miRNA expression or to employ small molecules to increase or inhibit miRNA function.14 Nevertheless, more efficient and selective in vivo delivery systems are needed to minimize the risk of unwanted side effects that can arise from a systemic delivery of miRNAs.14
Exposure in early development might influence the insurgence of diabetes and CV phenotypes later in life through persistent epigenetic changes, which may be inherited through mitotic and/or meiotic (‘transgenerational effect’) mechanisms.15–19 Indirect proofs demonstrated a strong correlation between children exposed to prenatal famine, low birth weight, and increased risk of Type 2 diabetes (T2D), hypertension and other CV diseases.15–19 According to the Barker hypothesis, the caloric restriction during pregnancy may permanently perturb crucial epigenetic-sensitive molecular networks associated with the normal foetus development leading to CV diseases later in life.16,19 Recently, a string correlation between maternal hypercholesterolaemia and hypermethylation of the sterol regulatory element-binding protein 2 (SREBP2) gene was observed in humans; however, the direct causality is difficult to infer from observational studies.17 Although the clinical implications for primary prevention are logical and intuitive in this field, the transgenerational effect in humans is still controversial and this issue should deserve further investigations.
Postnatal epigenetic sensors and intermediate phenotypes in prediction of diabetic cardiovascular complications
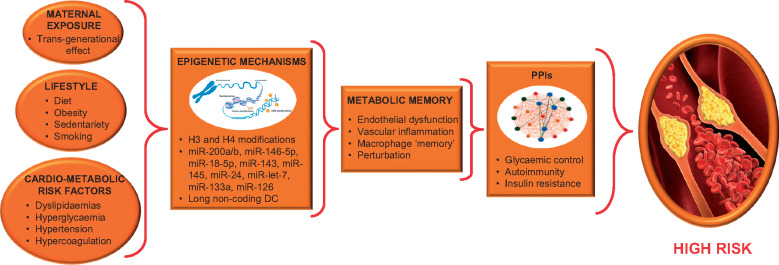
Our work has focused on epigenetic-sensitive mechanisms underlying key endophenotypes of diabetic vasculature, with much emphasis on macrovascular domains and coronary heart disease (CHD) (Figure 1). Furthermore, we reported the potential role of these epigenetic sensors as useful biomarkers in primary and secondary prevention of the CV risk in diabetic patients (Table 1). In order to highlight the increasing interest on epigenetics, we also included a brief list of ongoing or completed clinical trials but currently without any published results (Supplementary material online, Table S1).
Figure 1.
Epigenetic sensors, metabolic memory, and diabetes cardiovascular risk. Hyperglycaemia-induced epigenetic mechanisms may be transmitted through mitotic and/or meiotic (transgenerational effect) inheritance leading to detrimental changes in vasculature of diabetic patients. A persistent chromatin remodelling (metabolic memory) leads to perturbations of protein–protein interactions increasing the risk of coronary heart disease onset. CHD, coronary heart disease; MiRNAs, micro-RNAs; PPIs, protein–protein interactions.
Table 1.
Useful epigenetic-sensitive biomarkers in prevention of diabetes and its cardiovascular risk
| Epigenetic tag | Sample source | Effect | References |
|---|---|---|---|
| Primary prevention | |||
| DNA hypomethylation and histone 3 hyperacetylation | PBMCs | Increased oxidative stress | 20 |
| Down-regulation of miR-200a and miR-200b | HAECs | Increased endothelial inflammation | 21 |
| Down-regulation of miR-146a-5p | HAECs | Increased endothelial inflammation | 22 |
| Overexpression of miR-18a-5p | HAECs | Increased myocardial fibrosis | 23 |
| Down-regulation of miR-24 | Blood samples | Increased proinflammatory signalling | 24 |
| Hyperacetylation of H3K9 | Monocytes | Increased proinflammatory signalling | 25 |
| Hypermethylation of H3K4 | PBMCs | Increased endothelial inflammation | 26 |
| Secondary prevention | |||
| Down-regulation of miR-let-7 | Carotid plaque biopsy | Increased vascular inflammation | 27 |
| Down-regulation of miR-133a | LV samples | Increased autophagy and hypertrophy rate | 28 |
HAECs, human aortic endothelial cells; LV, left ventricular; MiR, micro-RNA; PBMCs, peripheral blood mononuclear cells.
Endothelial dysfunction and vascular inflammation
The endothelial monolayer is particularly susceptible to hyperglycaemia-induced injury leading to a complex cascade of pro-inflammatory and pro-oxidative signalling pathways associated with onset and progression of atherosclerotic lesions.29 A simultaneous DNA hypomethylation and histone 3 hyperacetylation occurred at level of p66 Src homologous-collagen homologue (p66Shc) gene promoter led to an overexpression of p66Shc protein in peripheral blood mononuclear cells increasing the oxidative stress in euglycaemic T2D patients with respect to controls.20 These molecular alterations may trigger the progression of oxidative stress-induced damages in the diabetic vasculature, suggesting novel predictive biomarkers and drug targets.20 Several miRNAs were involved in progression of endothelial dysfunction. For example, a down-regulation both of miR-200a and miR-200b induced up-regulation of O- linked N-acetylglucosamine transferase enzyme and then modification of O-linked N-acetylglucosamine proteins in human endothelial cells.21 These molecular changes were associated with hyperglycaemia-induced endothelial inflammation and diabetic complications.21 Furthermore, after treatment with high glucose concentration down-regulation of miR-146a-5p levels led to chronic inflammation by inducing higher levels of interleukin-1 receptor-associated kinase-1 (IRAK-1) gene expression in human endothelial cells.22 This evidence suggests that administration of miR-200a, miR-200b, and miR-146a-5p mimics might be useful therapeutic strategies to prevent inflammation and endothelial dysfunction in diabetic patients and their role should be further investigated in clinical studies. Moreover, increased levels of miR-18a-5p contributed to endothelial-mesenchymal transition phenotype by targeting the neurogenic locus notch homologue protein 2 (NOTCH2) signalling pathway in human endothelial cells under high glucose stimulation. This suggested that miR-18a-5p may act as a novel promising target for myocardial fibrosis in diabetic cardiomyopathy.23 Furthermore, levels of circulating miR-24 were lower in peripheral blood samples collected from T2D+/CHD+ patients as compared with CHD+ and control groups suggesting this molecule as a biomarker for predicting the onset of CHD in diabetic patients.24 In diabetic human carotid plaques, lower levels of miR-let-7 were correlated to chronic inflammation.27 Interestingly, ex vivo administration of miR-let-7 mimics to human carotid plaques was able to module the inflammatory phenotype indicating a putative therapeutic strategy to prevent growth and rupture of atherosclerotic plaques in diabetic patients.27 Moreover, a downregulation of miR-133a was detected in cardiac cells isolated from left ventricle of diabetic patients undergoing left ventricular assist device implantation with respect to controls.28 This suggested a useful clinical biomarker of cardiac autophagy and hypertrophy in diabetic patients complicated by heart failure.28
Macrophage ‘memory’
A genome-wide analysis revealed higher levels of acetylation at lysine 9 of histone 3 located to several gene promoters related to the nuclear factor-kB (NF-KB) inflammatory pathway and other diabetes complications in monocytes isolated from T1D patients with respect to control group.25 These molecular signatures suggested that a global hyperacetylation may be a possible epigenetic explanation for metabolic memory in humans. Moreover, increased levels of mono-methylation at histone 3 (H3K4m1), mediated by Set7 DNA methyltransferase enzyme, led to up-regulation of RELA proto-oncogene in peripheral blood mononuclear cells isolated from T2D patients respect with controls.26 This evidence suggested another aberrant epigenetic change contributing to vascular dysfunction in T2D patients.26 Taken together, these results suggested that specific histone 3 modifications may be possible mechanisms of metabolic memory and then novel therapeutic approaches to prevent atherosclerosis in diabetes.
Epitranscriptomics
The role of environmental-induced chemical modifications occurring in regulatory RNAs (epitrascriptome) in the prevention of CV risk in diabetes is currently little investigated. Generally, non-coding RNAs are regulators of gene expression acting at post-transcriptional level; however, heterogeneous chemical modifications occurring in these molecules may generate an additional layer of gene expression control known as ‘epitrascriptome’.30 Many chemical perturbations in transfer RNA or dietary factors may induce a fragmentation of these molecules that seems to be associated with damage of pancreatic β-cell damage in diabetes.31 Moreover, the content of the N6-methyladenosine (m6A) in mRNAs, referring to the methylation of the adenosine base at the nitrogen-6 position, was significantly lower in peripheral blood samples isolated from T2D patients with respect to healthy controls.32 Additionally, the lower m6A content in mRNA was correlated with higher expression of the fat mass- and obesity-associated demethylase enzyme suggesting a putative mechanistic link and a useful biomarker of T2D onset.33
Strategies for longitudinal analyses and modelling of epigenetic trajectories in diabetes cardiovascular risk
Since the epigenome largely varies over time, the possibility of studying molecular changes at different time points may be an added value to our understanding of their putative clinical relevance. Indeed, in this way, we may identify the genomic sites and circulating molecules with the largest associations with the environmental changes with respect to analyses at one point in time (cross-sectional studies).33
For example, lower plasma levels of miR-126 were detected in the prospective population-based Bruneck study including T2D patients (n = 822) with respect to age- and sex-matched controls.34 In detail, lower levels of miR-126 were revealed in endothelial apoptotic bodies associated with impaired angiogenic signalling.34 Importantly, miR-126 plasma variations were detected prior to insurgence of T2D clinical manifestations suggesting its useful clinical role as predictive biomarker.34 Successively, the endothelial-enriched miR-126 also showed a positive association with subsequent myocardial infarction events in the same population study, suggesting a useful biomarker in secondary prevention of diabetic patients.35 Thus, future studies will be needed to address whether platelet-derived endothelial-enriched miR-126 may serve as novel biomarker for clinical decision-making.
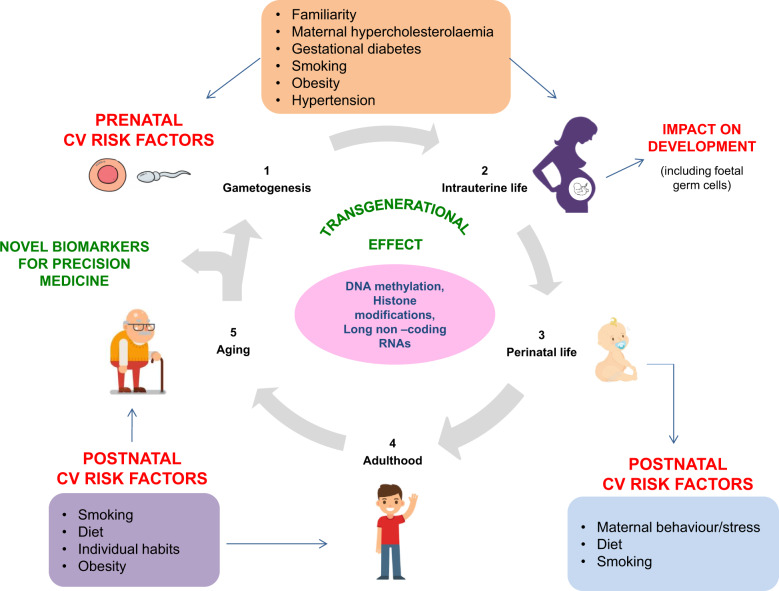
Since the maternal intrauterine environment may shape the individual long-term susceptibility to diabetes, we remark that future prospective clinical studies should map the individual epigenetic signatures in high-risk families at multiple time points across several generations (transgenerational effect).15–19 This paradigm may provide reliable and stable biomarkers to identify at-risk subjects before the development of clinical signs of vascular damages, with important implications for improving primary prevention (Figure 2).
Figure 2.
An integrated approach to study epigenetic mechanisms underlying diabetes cardiovascular risk over time. Epigenetics changes may be inherited from one generation to the next, thus affecting the individual susceptibility to develop diabetes and its cardiovascular complications. Effects of prenatal exposures and postnatal exposures should be investigated in humans, in order to clarify earlier pathogenic determinants in the diabetic vasculature. T1D, diabetes Type 1; T2D, diabetes Type 2.
Epidrugs: the road ahead of clinical trials for prevention of diabetes cardiovascular risk
In Table 2, we summarized the list of ongoing and completed clinical trials (https://clinicaltrials.gov) with published results in the last 10 years.
Table 2.
Some epidrugs in clinical trials to prevent diabetes cardiovascular risk
| Epidrug | Conditions | Effect | Phase/status | NCT |
|---|---|---|---|---|
| Primary prevention | ||||
| Monotherapy | ||||
| Metformin | Obesity, T2D | To assess the efficacy in reducing BMI. | Phase 4/completed | NCT00934570 36 |
| Combination therapy | ||||
| Metformin plus insulin | T1D | To test the efficacy in reducing atherosclerosis. | Phase 3/completed | NCT01483560 37 |
| Fenofibrate plus statins | Atherosclerosis, CHD, T2D | To prevent major macrovascular complications. | Phase 3/completed | NCT00000620 38 |
| Secondary prevention | ||||
| Monotherapy | ||||
| Metformin or glipizide | T2D, CHD | To test if metformin may reduce the risk of recurrent CV events. | Phase 4/completed | NCT00513630 39 |
| Combination therapy | ||||
| Metformin plus insulin analogues | T2D, atherosclerosis | To prevent progression of carotid IMT. | Phase 4/completed | NCT00657943 40 |
| Metformin plus vildagliptin | T2D, CHD | To reduce inflammation. | Phase 4/completed | NCT01604213 41 |
BMI, body mass index; CHD, coronary heart disease; CV, cardiovascular; IMT, intima-media thickness; NA, not applicable; NCT, ClinicalTrials.gov Identifier number; T1D, type 1 diabetes; T2D, Type 2 diabetes.
Primary prevention
Metformin is the first-line oral drug for the initial treatment of T2D patients.2 Metformin belongs to HDAC activator class; it acts as agonist of the silent information regulator 1 (SIRT1) enzyme involved in reduction of insulin resistance, mitochondrial dysfunction, and oxidative stress reporting a key role in prevention of heart failure.6,42 A combination therapy based on lifestyle changes (diet and exercise) and metformin declined the body mass index at 1 year of initial observation both in obese children and adolescents with respect to singular treatments.36 Moreover, 3 years of treatment with metformin combined with titrated insulin therapy reduced atherosclerosis in T1D adult patients but did not improve glycaemic control in these patients suggesting that this approach could have a wider role in CV risk management.37
Fenofibrate, as HDAC activator, is an antihypertriglycerid agent useful for treatment of hypertriglyceridemia. At molecular level, it acts as agonist of peroxisome proliferator-activated receptor alpha protein that is involved in regulation of glucose metabolism and insulin sensitivity.43 A clinical trial tested the efficacy of fenofibrate to gain euglycaemia state and blood pressure control in T2D patients treated with statins.38 As HDAC inhibitor, statins act as modulators of the 3-hydroxy-3-methylglutaryl coenzyme A reductase enzyme representing the gold standard cholesterol-lowering agent treatment of diabetic patients.2,44 This trial demonstrated that fenofibrate therapy may reduce the CV risk in these patients; however, a larger investigation is needed to confirm these findings.38
Secondary prevention
By comparing the risk of recurrent CV events in T2D+/CHD+ patients, it raised that 3 years of treatment with metformin strongly reduced vascular dysfunction in high-risk patients compared with glipizide therapy.39 Moreover, 18 months of treatment with metformin combined with insulin analogue did not reduce the progression of mean carotid intima-media thickness in T2D patients.40 Instead, a combination of vildagliptin plus metformin led to a significant anti-inflammatory phenotype during follow-up in T2D+/CHD+ patients, owing to suppression of the interleukin (IL) IL-1B activity.41 Apabetalone (or RVX 208) is an inhibitor of bromodomain extra terminal (BET) proteins, mainly BED4, which contain two bromodomains interacting with acetylated lysines of histone tails.45 Experimental evidence reported that when apabetalone binds to BRD4 some deregulated molecular pathways involved in cholesterol metabolism and inflammation are restored in humans.45 In detail, administration of RVX 208 could increase plasma high-density lipoprotein levels, reduce oral glucose absorption and endogenous glucose production in prediabetics with respect to controls, suggesting a protective role against T2D onset.45 To date, apabetalone is in Phase III ongoing trial (NCT02586155) assessing whether BET inhibition may prevent the risk of CV events in T2D+-CHD+ patients.
Taken together, these results will open new perspectives in personalized therapy of diabetic patients predicting which patients may benefit or not from a specific monotherapy or combination therapy.
A new challenge for personalized therapy
Several national biobanks (e.g. UK biobank) were developed in useful quantitative polygenic score platforms able to diagnose or predict the risk of diabetes and CV disease onset.3,46,47 Through these platforms and generation of huge amount of big data, advanced network-based algorithms may aid the prioritization of genes and molecules with a functional relevance for preventing and treating the CV risk in diabetics.46,48–50 These algorithms enable a quantitative analysis of the network topological properties providing a graphical map where human diseases are represented with localized perturbation within a specific module of the cellular interactome.48 A biological network is a set of nodes and edges, in which the nodes are linked if there is a functional or physical interaction between them.48 Protein–protein interactions (PPIs), regulatory and co-expression networks can reflect crucial nodes (proteins, genes, etc.) or disease modules (a subgroup of nodes) able to stratify the risk and/or predict drug responses at individual level.48 In PPI networks, including Gene Prioritizing Approach using Network Distance Analyses (GenePanda) and Degree-Aware Disease Gene Prioritization (DIAMOnD), the nodes are proteins that are linked to each other by physical interactions; in regulatory networks, including Passing Attributes between Networks for Data Assimilation (PANDA), the direct links represent regulatory relationships between a transcription factor and a gene whereas in co-expression networks, including Weighted Correlation Network Analysis (WGCNA), gene pairs with similar co-expression profiles are linked in the same disease module.48 We discuss two examples of network tools to identify genetic/epigenetic nodes with a putative clinical impact for prevention and therapy of diabetes and its CV risk.49,50 By analysing big data extracted from human pancreatic islets, some authors constructed a tissue-specific gene regulatory network.49 In detail, an enrichment of variants in the nuclear factor of activated T cells 4 (NFATC4) gene was observed in the islet regulatory network of T2D patients suggesting a crucial role in pathogenesis of disease.49 Moreover, a PPI network revealed that a differential expression of some miRNAs could play a crucial role in pathogenesis of metabolic disorders in T2D patients.50 By results, a modification of hsa-miR-4687-3p levels in samples of epicardial adipose tissues may play a crucial role in discriminating CV events in T2D+/CHD+ patients as compared to controls, suggesting an additional useful diagnostic biomarker.50 Despite this basic findings, more larger prospective and independent population-based studies should be performed to validate and translate in clinic these preliminary results.
Integrating machine learning with protein–protein interactions to predict novel biomarkers in diabetes and its cardiovascular risk
Machine learning algorithms mainly include artificial neural networks, Bayesian networks, support vector machines, and decision trees, which are widely applied in clinical research for the development of predictive models by extracting key variables from large clinical datasets.51 A large clinical trial (NCT00005487) tested machine learning algorithms combined with deep phenotyping and provided a model able to predict six CV outcomes in asymptomatic subjects from the Multi-Ethnic Study of Atherosclerosis (MESA) study.52 From preliminary results, these methods may lead to greater accuracy in prediction of CV diseases but an appropriate level of validation is still needed before translating these models in the everyday clinical practice.
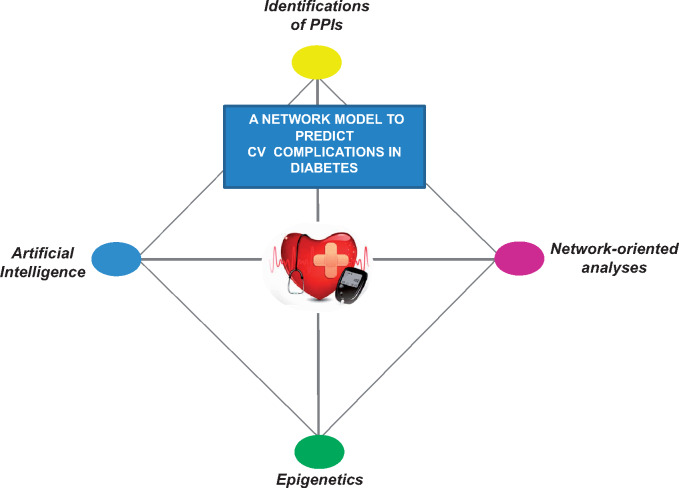
New opportunities may arise from application of machine learning algorithms into predicting PPI networks.53 For this aim, a novel machine learning tool, named DPPI, has been developed to accurately predict protein interactions from DNA sequence information alone (https://github.com/hashemifar/DPPI/).54 We remark that an integrated approach combining machine learning algorithms and epigenomics datasets may extend our knowledge of diabetic interactome revealing additional useful biomarkers for the prediction of CV risk in diabetes (Figure 3).
Figure 3.
An integrative approach to identify protein–protein interactions by combining network-based algorithms, machine learning, and epigenetics. The strong predictive power of machine learning and network-based algorithms should be combined with epigenomic datasets of thousands of patients in order to clarify how, when, and how molecular alterations may perturb the diabetic interactome. CV, cardiovascular; PPIs, protein–protein interactions.
Machine learning and tomography
Although in this manuscript we mainly focused on diabetes and its macrovascular complications, we discuss the putative clinical benefit from combining machine learning and tomography in evaluating vascular lesions both in micro- and macro-domains.55 In a recent study on 10 030 T2D patients and suspected CHD, 25 clinical parameters and 44 imaging measures were elaborated in machine learning algorithms.56 This study provided a model able to predict 5-year all-cause mortality better than clinical or coronary computed tomography angiography variables alone.55 Similar evidence arose from application of machine learning to singular images derived from the optical coherence tomography angiography technique. As a result, this strategy provided an accurate dynamic map of the retinal vascular flow.57 These recent advances are demonstrating that development of machine learning algorithms may enable huge opportunities for prevention and diagnosis of diabetes and its CV complications increasing chances for personalized therapy.
Conclusions
A plethora of epigenetic-sensitive changes, mainly miR-126, was identified in diabetic patients suggesting a pathogenic role relevant to progression towards long-term vascular complications in different human tissues. Importantly, these epigenetic signatures may be inherited across several generations (transgenerational effect). This concept strengthens their putative clinical role as predictive biomarkers of CV complications. Furthermore, the plasticity of epigenome allows the modulation of genome by environmental changes. As consequence, lifestyle modulation combined with epidrugs, such as metformin, statins, and fenofibrate may help to reach personalized therapy of diabetic patients. Despite the well-established difference between man and woman in CV diseases, the majority of studies do not show their data stratified by gender. Moreover, we are now unable to fully understand what is the impact of epigenetics for T1 vs. T2 diabetes as well as for rarer monogenic forms like maturity-onset diabetes of the young (MODY).58
Future perspectives
Despite increasing development of risk prediction models and the solid guidelines approach of reductionist practice, physicians still fail into predicting the onset of diabetes and progression of its micro- and macrovascular complications. Epigenomics offers precious information on the specific spatio-temporal state of cells and tissues increasing the opportunity to identify specific molecular changes contributing to identification of the individual CV risk in diabetic patients. Owing to the high mortality rate, it is mandatory to search innovative molecular fingerprints helping us to identify patients at high-risk to develop micro- and macrovascular diabetes complications. DNA methylation and miRNAs are major determinants involved in diabetic interactome. Moreover, DNA methylation is not only influenced by genetic factors, whereas large differences in blood-based methylation profile are founded when the population study is gender-stratified suggesting the need for longitudinal prospective studies to support the robustness of these associations.59 In this regard, identification of epigenetic interactions between miRNAs and DNA methylation associated with gene expression may aid to advance our knowledge on molecular basis underlying the CV risk in diabetic patients providing putative useful biomarkers.
Recently, we have designed the ‘Perturbation of Interactome through micro-RNA and Methylome analysis In Diabetes Endophenotypes: the PIRAMIDE pathogenic clinical study design’ (NCT03792607).60 This ongoing pilot study combines epigenetic interactions and network-based algorithms in order to provide useful biomarkers predicting the onset of macrovascular complications in T2D patients.60 The similarity of our approach was present in two studies analysing the interaction of epigenetic mechanisms in diabetic patients complicated with CHD and their putative clinical role as biomarkers or drug targets.48,49 Moreover, it is crucial to investigate how epigenetic modifications may affect the individual response to pharmacological and non-pharmacological treatments, including lifestyle changes such as dietary modifications, exercise, avoiding stress, and minimizing alcohol consumption. Combining clinical evaluation and genetic data with the increasing epigenetic information is the most fruitful way to reach personalized therapy for diabetes care.5,15,50,61
Supplementary material
Supplementary material is available at European Heart Journal – Cardiovascular Pharmacotherapy online.
Funding
This work was supported by PRIN2017F8ZB89 from Italian Ministry of Research (PI Prof Napoli). G.B. is a PhD student of Translational Medicine awarded ESC Congress 2019 and she is supported by Educational Grant from the University of Campania “Luigi Vanvitelli”, Naples, Italy.
Conflict of interest: none declared.
Supplementary Material
References
- 1. Domingueti CP, Dusse LM, Carvalho M, de Sousa LP, Gomes KB, Fernandes AP.. Diabetes mellitus: the linkage between oxidative stress, inflammation, hypercoagulability and vascular complications. J Diabetes Complications 2016;30:738–745. [DOI] [PubMed] [Google Scholar]
- 2. Cosentino F, Grant PJ, Aboyans V, Bailey CJ, Ceriello A, Delgado V, Federici M, Filippatos G, Grobbee DE, Hansen TB, Huikuri HV, Johansson I, Jüni P, Lettino M, Marx N, Mellbin LG, Östgren CJ, Rocca B, Roffi M, Sattar N, Seferović PM, Sousa-Uva M, Valensi P, Wheeler DC; ESC Scientific Document Group. 2019 ESC Guidelines on diabetes, pre-diabetes, and cardiovascular diseases developed in collaboration with the EASD. Eur Heart J 2019;doi: 10.1093/eurheartj/ehz486. [DOI] [PubMed] [Google Scholar]
- 3. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, Kathiresan S.. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hallén J, Sreeharan N.. Development of triglyceride-lowering drugs to address residual cardiovascular risk: strategic and clinical considerations. Eur Heart J Cardiovasc Pharmacother 2018;4:237–242. [DOI] [PubMed] [Google Scholar]
- 5. Keating ST, Plutzky J, El-Osta A.. Epigenetic changes in diabetes and cardiovascular risk. Circ Res 2016;118:1706–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sommese L, Benincasa G, Lanza M, Sorriento A, Schiano C, Lucchese R, Alfano R, Nicoletti GF, Napoli C.. Novel epigenetic-sensitive clinical challenges both in type 1 and type 2 diabetes. J Diabetes Complications 2018;32:1076–1084. [DOI] [PubMed] [Google Scholar]
- 7. Karlson BW, Wiklund O, Palmer MK, Nicholls SJ, Lundman P, Barter PJ.. Variability of low-density lipoprotein cholesterol response with different doses of atorvastatin, rosuvastatin, and simvastatin: results from VOYAGER. Eur Heart J Cardiovasc Pharmacother 2016;2:212–217. [DOI] [PubMed] [Google Scholar]
- 8. Loscalzo J, Handy DE.. Epigenetic modifications: basic mechanisms and role in cardiovascular disease (2013 Grover Conference series). Pulm Circ 2014;4:169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van der Wijst MG, Venkiteswaran M, Chen H, Xu GL, Plösch T, Rots MG.. Local chromatin microenvironment determines DNMT activity: from DNA methyltransferase to DNA demethylase or DNA dehydroxymethylase. Epigenetics 2015;10:671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schiano C, Vietri MT, Grimaldi V, Picascia A, De Pascale MR, Napoli C.. Epigenetic-related therapeutic challenges in cardiovascular disease. Trends Pharmacol Sci 2015;36:226–235. [DOI] [PubMed] [Google Scholar]
- 11. Fritz KS. Chemical acetylation and deacetylation. Methods Mol Biol 2013;1077:191–201. [DOI] [PubMed] [Google Scholar]
- 12. Carthew RW, Sontheimer EJ.. Origins and mechanisms of miRNAs and siRNAs. Cell 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Rooij E, Purcell AL, Levin AA.. Developing microRNA therapeutics. Circ Res 2012;110:496–507. [DOI] [PubMed] [Google Scholar]
- 14. Tahamtan A, Teymoori-Rad M, Nakstad B, Salimi V.. Anti-inflammatory microRNAs and their potential for inflammatory diseases treatment. Front Immunol 2018;9:1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Napoli C, Crudele V, Soricelli A, Al-Omran M, Vitale N, Infante T, Mancini FP.. Primary prevention of atherosclerosis: a clinical challenge for the reversal of epigenetic mechanisms? Circulation 2012;125:2363–2373. [DOI] [PubMed] [Google Scholar]
- 16. Napoli C, Infante T, Casamassimi A.. Maternal-foetal epigenetic interactions in the beginning of cardiovascular damage. Cardiovasc Res 2011;92:367–374. [DOI] [PubMed] [Google Scholar]
- 17. de Nigris F, Cacciatore F, Mancini FP, Vitale DF, Mansueto G, D’Armiento FP, Schiano C, Soricelli A, Napoli C.. Epigenetic hallmarks of fetal early atherosclerotic lesions in humans. JAMA Cardiol 2018;3:1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oluwagbemigun K, Buyken AE, Alexy U, Schmid M, Herder C, Nöthlings U.. Developmental trajectories of body mass index from childhood into late adolescence and subsequent late adolescence-young adulthood cardiometabolic risk markers. Cardiovasc Diabetol 2019;18:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zanetti D, Tikkanen E, Gustafsson S, Priest JR, Burgess S, Ingelsson E.. Type 2 diabetes mellitus, and cardiovascular disease: addressing the Barker hypothesis with Mendelian randomization. Circ Genom Precis Med 2018;11:e002054.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costantino S, Paneni F, Battista R, Castello L, Capretti G, Chiandotto S, Tanese L, Russo G, Pitocco D, Lanza GA, Volpe M, Lüscher TF, Cosentino F.. Impact of glycemic variability on chromatin remodeling, oxidative stress, and endothelial dysfunction in patients with type 2 diabetes and with target HbA(1c) levels. Diabetes 2017;66:2472–2482. [DOI] [PubMed] [Google Scholar]
- 21. Lo WY, Yang WK, Peng CT, Pai WY, Wang HJ.. MicroRNA-200a/200b modulate high glucose-induced endothelial inflammation by targeting O-linked N-acetylglucosamine transferase expression. Front Physiol 2018;9:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lo WY, Peng CT, Wang HJ.. MicroRNA-146a-5p mediates high glucose-induced endothelial inflammation via targeting interleukin-1 receptor-associated kinase 1 expression. Front Physiol 2017;8:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geng H, Guan J.. MiR-18a-5p inhibits endothelial-mesenchymal transition and cardiac fibrosis through the Notch2 pathway. Biochem Biophys Res Commun 2017;49:329–336. [DOI] [PubMed] [Google Scholar]
- 24. Deng X, Liu Y, Luo M, Wu J, Ma R, Wan Q, Wu J.. Circulating miRNA-24 and its target YKL-40 as potential biomarkers in patients with coronary heart disease and type 2 diabetes mellitus. Oncotarget 2017;8:63038–63046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miao F, Chen Z, Genuth S, Paterson A, Zhang L, Wu X, Li SM, Cleary P, Riggs A, Harlan DM, Lorenzi G, Kolterman O, Sun W, Lachin JM, Natarajan R; DCCT/EDIC Research Group. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014;63:1748–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paneni F, Costantino S, Battista R, Castello L, Capretti G, Chiandotto S, Scavone G, Villano A, Pitocco D, Lanza G, Volpe M, Lüscher TF, Cosentino F.. Adverse epigenetic signatures by histone methyltransferase Set7 contribute to vascular dysfunction in patients with type 2 diabetes mellitus. Circ Cardiovasc Genet 2015;8:150–158. [DOI] [PubMed] [Google Scholar]
- 27. Brennan E, Wang B, McClelland A, Mohan M, Marai M, Beuscart O, Derouiche S, Gray S, Pickering R, Tikellis C, de Gaetano M, Barry M, Belton O, Ali-Shah ST, Guiry P, Jandeleit-Dahm KAM, Cooper ME, Godson C, Kantharidis P.. Protective effect of let-7 miRNA family in regulating inflammation in diabetes-associated atherosclerosis. Diabetes 2017;66:2266–2277. [DOI] [PubMed] [Google Scholar]
- 28. Nandi SS, Duryee MJ, Shahshahan HR, Thiele GM, Anderson DR, Mishra PK.. Induction of autophagy markers is associated with attenuation of miR-133a in diabetic heart failure patients undergoing mechanical unloading. Am J Transl Res 2015;7:683–696. [PMC free article] [PubMed] [Google Scholar]
- 29. Tang R, Gao M, Wu M, Liu H, Zhang X, Liu B.. High glucose mediates endothelial-to-chondrocyte transition in human aortic endothelial cells. Cardiovasc Diabetol 2012;11:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roundtree IA, Evans ME, Pan T, He C.. Dynamic RNA modifications in gene expression regulation. Cell 2017;169:1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cosentino C, Cnop M, Igoillo-Esteve M.. The tRNA epitranscriptome and diabetes: emergence of tRNA hypomodifications as a cause of pancreatic β-cell failure. Endocrinology 2019;160:1262.. [DOI] [PubMed] [Google Scholar]
- 32. Shen F, Huang W, Huang JT, Xiong J, Yang Y, Wu K, Jia GF, Chen J, Feng YQ, Yuan BF, Liu SM.. Decreased N(6)-methyladenosine in peripheral blood RNA from diabetic patients is associated with FTO expression rather than ALKBH5. J Clin Endocrinol Metab 2015;100:148–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Napoli C, Benincasa G, Loscalzo J.. Epigenetic inheritance underlying pulmonary arterial hypertension. Arterioscler Thromb Vasc Biol 2019;39:653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M.. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010;107:810–817. [DOI] [PubMed] [Google Scholar]
- 35. Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S, Mayr M.. Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 2012;60:290–299. [DOI] [PubMed] [Google Scholar]
- 36. Wilson AJ, Prapavessis H, Jung ME, Cramp AG, Vascotto J, Lenhardt L, Shoemaker JK, Watson M, Robinson T, Clarson CL.. Lifestyle modification and metformin as long-term treatment options for obese adolescents: study protocol. BMC Public Health 2009;9:434.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrie JR, Chaturvedi N, Ford I, Brouwers MCGJ, Greenlaw N, Tillin T, Hramiak I, Hughes AD, Jenkins AJ, Klein BEK, Klein R, Ooi TC, Rossing P, Stehouwer CDA, Sattar N, Colhoun HM, Nickerson H, Lou O, Dutta S, Haw J, Anderson C, Kean S, Thomson E, Gillespie L, Gibb J, Greenlaw N, Keech A, Jenkins A, March K, Williams S, Coady E, Bots M, Dreyer J, Jan T, Sheffy K, Lusky R, Peleg S, Shore A, Carty D, Donnan P, Witham M, Adler A, Lonn E, Rauchhaus P, Lindsay R, Brouwers M, Van-Melckebeke J, Gillespie L, Hamill T, Cuthbertson L, Murray A, Jolly L, Miller E, Hair J, Bell A, Carmichael S, Douglas E, Surtees P, Dinnett E, Allan J, Watson C, McLaughlin M, Brindley G, Smillie E, Motherwell D, MacDonald S, Ellis P, Stuart D, Travers M, Brearley S, Greig L, Colman P, Nankervis A, Forulanos S, West D, Vaughan S, Bjorasen M, Donlan J, Vrazas J, O'Neal D, Horsburgh J, Pater H, Kent S, Twigg S, Fulcher G, Denner R, Piotrowicz A, Januszewski A, Coy A, Paul T, McDonald C, Tereschyn S, Schmidt N, Weingert M, Heard H, Burke S, Ooi TC, Lochnan H, Sorisky A, Keely E, Malcolm J, Maranger J, Favreau C, Petherick S, Boles K, Rossing P, Hansen TW, Lund S, Hemmingsen B, Thorogood N, Green K, Robinson T, Abouglilia K, Nayman D, Miller C, Warren R, Aizawa K, Balasubramani M, Toth S, Harvey K, Birch G, Atkin S, Sathyapalan T, James A, Javed Z, Wilding J, Martin B, Birch S, Wilcox A, Watson N, Oliver N, Jugnee N, Rutter M, Turgut T, Shaju A, Yau S, Subin S, Walker M, Wake D, Miller C, Millward A, Chong P, Hibbert M, George J, Schaper N, Pinxt J, Op Het Roodt J, Phillips S, Murray L, Sleigh L, Collier A, Sit LE, Allan K, Cook J, Campbell K, Hodge L, Leese G, Reekie G, Jaap A, Sudworth A, White A, McKnight J, Steven L, McKay G, Llano A, Currie G, Lennon E, Johnstone J, Shields K.. Cardiovascular and metabolic effects of metformin in patients with type 1 diabetes (REMOVAL): a double-blind, randomised, placebo-controlled trial. Lancet Diabetes Endocrinol 2017;5:597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Elam MB, Ginsberg HN, Lovato LC, Corson M, Largay J, Leiter LA, Lopez C, O’Connor PJ, Sweeney ME, Weiss D, Friedewald WT, Buse JB, Gerstein HC, Probstfield J, Grimm R, Ismail-Beigi F, Goff DC, Fleg JL, Rosenberg Y, Byington RP; ACCORDION Study Investigators . Association of fenofibrate therapy with long-term cardiovascular risk in statin-treated patients with type 2 diabetes. JAMA Cardiol 2017;2:370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong J, Zhang Y, Lai S, Lv A, Su Q, Dong Y, Zhou Z, Tang W, Zhao J, Cui L, Zou D, Wang D, Li H, Liu C, Wu G, Shen J, Zhu D, Wang W, Shen W, Ning G; SPREAD-DIMCAD Investigators. Effects of metformin versus glipizide on cardiovascular outcomes in patients with type 2 diabetes and coronary artery disease. Diabetes Care 2013;36:1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lundby-Christensen L, Vaag A, Tarnow L, Almdal TP, Lund SS, Wetterslev J, Gluud C, Boesgaard TW, Wiinberg N, Perrild H, Krarup T, Snorgaard O, Gade-Rasmussen B, Thorsteinsson B, Røder M, Mathiesen ER, Jensen T, Vestergaard H, Hedetoft C, Breum L, Duun E, Sneppen SB, Pedersen O, Hemmingsen B, Carstensen B, Madsbad S.. Effects of biphasic, basal-bolus or basal insulin analogue treatments on carotid intima-media thickness in patients with type 2 diabetes mellitus: the randomised Copenhagen Insulin and Metformin Therapy (CIMT) trial. BMJ Open 2016;6:e008377.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Younis A, Eskenazi D, Goldkorn R, Leor J, Naftali-Shani N, Fisman EZ, Tenenbaum A, Goldenberg I, Klempfner R.. The addition of vildagliptin to metformin prevents the elevation of interleukin 1B in patients with type 2 diabetes and coronary artery disease: a prospective, randomized, open-label study. Cardiovasc Diabetol 2017;16:69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cittadini A, Napoli R, Monti MG, Rea D, Longobardi S, Netti PA, Walser M, Samà M, Aimaretti G, Isgaard J, Saccà L.. Metformin prevents the development of chronic heart failure in the SHHF rat model. Diabetes 2012;61:944–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scognamiglio M, Costa D, Sorriento A, Napoli C.. Current therapy and nutraceuticals for the treatment of patients with dyslipidemias. Curr Pharm Des 2019;25:85–95. [DOI] [PubMed] [Google Scholar]
- 44. Vonbank A, Drexel H, Agewall S, Lewis BS, Dopheide JF, Kjeldsen K, Ceconi C, Savarese G, Rosano G, Wassmann S, Niessner A, Schmidt TA, Saely CH, Baumgartner I, Tamargo J.. Reasons for disparity in statin adherence rates between clinical trials and real-world observations: a review. Eur Heart J Cardiovasc Pharmacother 2019;5:36.. [DOI] [PubMed] [Google Scholar]
- 45. Nicholls SJ, Ray KK, Johansson JO, Gordon A, Sweeney M, Halliday C, Kulikowski E, Wong N, Kim SW, Schwartz GG.. Selective BET protein inhibition with apabetalone and cardiovascular events: a pooled analysis of trials in patients with coronary artery disease. Am J Cardiovasc Drugs 2018;18:109–115. [DOI] [PubMed] [Google Scholar]
- 46. Leopold JA, Loscalzo J.. Emerging role of precision medicine in cardiovascular disease. Circ Res 2018;122:1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, Sterling DG, Williams SA.. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 2016;315:2532–2541. [DOI] [PubMed] [Google Scholar]
- 48. Barabási AL, Gulbahce N, Loscalzo J.. Network medicine: a network-based approach to human disease. Nat Rev Genet 2011;12:56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sharma A, Halu A, Decano JL, Padi M, Liu YY, Prasad RB, Fadista J, Santolini M, Menche J, Weiss ST, Vidal M, Silverman EK, Aikawa M, Barabási AL, Groop L, Loscalzo J.. Controllability in an islet specific regulatory network identifies the transcriptional factor NFATC4, which regulates type 2 diabetes associated genes. NPJ Syst Biol Appl 2018;4:25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu Y, Fu W, Lu M, Huai S, Song Y, Wei Y.. Role of miRNAs in epicardial adipose tissue in CAD patients with T2DM. Biomed Res Int 2016;2016:1629236.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Deo RC. Machine learning in medicine. Circulation 2015;132:1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Hundley WG, McClelland R, Gomes AS, Folsom AR, Shea S, Guallar E, Bluemke DA, Lima J.. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res 2017;121:1092–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang M, Su Q, Lu Y, Zhao M, Niu B.. Application of machine learning approaches for protein-protein interactions prediction. Med Chem 2017;13:506–514. [DOI] [PubMed] [Google Scholar]
- 54. Hashemifar S, Neyshabur B, Khan AA, Xu J.. Predicting protein-protein interactions through sequence-based deep learning. Bioinformatics 2018;1:802–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Infante T, Forte E, Schiano C, Punzo B, Cademartiri F, Cavaliere C, Salvatore M, Napoli C.. Evidence of association of circulating epigenetic-sensitive biomarkers with suspected coronary heart disease evaluated by cardiac computed tomography. PLoS One 2019;14:e0210909.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Motwani M, Dey D, Berman DS, Germano G, Achenbach S, Al-Mallah MH, Andreini D, Budoff MJ, Cademartiri F, Callister TQ, Chang HJ, Chinnaiyan K, Chow BJ, Cury RC, Delago A, Gomez M, Gransar H, Hadamitzky M, Hausleiter J, Hindoyan N, Feuchtner G, Kaufmann PA, Kim YJ, Leipsic J, Lin FY, Maffei E, Marques H, Pontone G, Raff G, Rubinshtein R, Shaw LJ, Stehli J, Villines TC, Dunning A, Min JK, Slomka PJ.. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J 2017;38:500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee CS, Tyring AJ, Wu Y, Xiao S, Rokem AS, DeRuyter NP, Zhang Q, Tufail A, Wang RK, Lee AY.. Generating retinal flow maps from structural optical coherence tomography with artificial intelligence. Sci Rep 2019;9:5694.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tallapragada DS, Bhaskar S, Chandak GR.. New insights from monogenic diabetes for “common” type 2 diabetes. Front Genet 2015;6:251.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Willmer T, Johnson R, Louw J, Pheiffer C.. Blood-Based DNA methylation biomarkers for type 2 diabetes: potential for clinical applications. Front Endocrinol (Lausanne) 2018;9:744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Benincasa G, Marfella R, Schiano C, Napoli C.. Perturbation of Interactome through micro-RNA and Methylome analysis In Diabetes Endophenotypes: the PIRAMIDE pathogenic clinical study design. Int J Clin Trials 2019;6:117–121. [Google Scholar]
- 61. Trignano E, Fallico N, Chen HC, Faenza M, Bolognini A, Armenti A, Santanelli Di Pompeo F, Rubino C, Campus GV.. Evaluation of peripheral microcirculation improvement of foot after tarsal tunnel release in diabetic patients by transcutaneous oximetry. Microsurgery 2016;36:37–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.