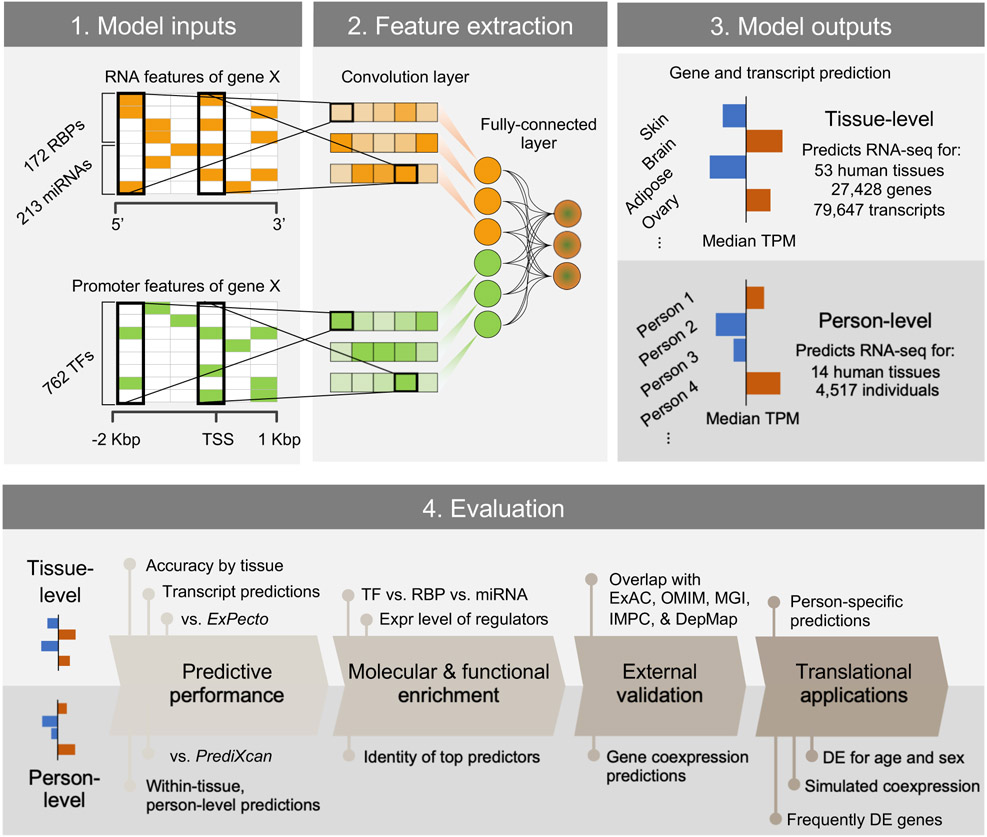
Figure 1.
Overview of building and evaluating the DEcode transcriptome prediction model. DEcode takes the promoter features and the mRNA features for each gene/transcript as inputs and outputs its expression levels under various conditions. We conducted a series of evaluations to show that DEcode defines major principles in gene regulation in arbitrary gene expression data. This capacity is strongly supported on a comparative basis to alternative methods, and on an absolute basis across diverse applications, which include, through predictions of transcript-usage, person-specific gene expression, frequently DE genes of multiple external disease-related gene sets.