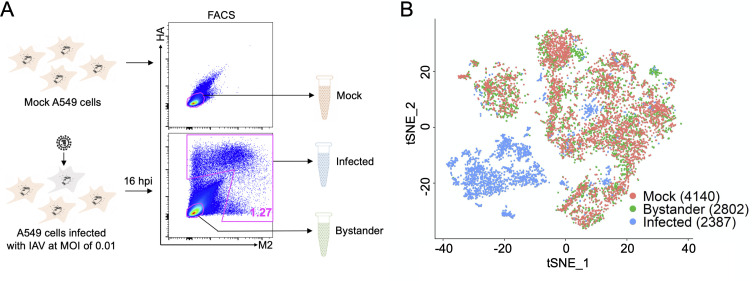
Fig 1. Generation of viral and host transcriptional data for thousands of singly infected cells.
(A) Schematic depicting our strategy for generating single cell RNAseq libraries from thousands of cells infected at low MOI. In brief, we infect A549 cells with Cal07 or Perth09 at MOI = 0.01 to ensure that infected cells are infected with a single virion. We then block secondary spread with NH4Cl treatment to make sure infection timing is uniform across all infected cells. Finally, we sort “infected” and “bystander” cells based on surface expression of HA and/or M2 and immediately generate single cell RNAseq libraries from these sorted cell populations using the 10X Chromium device. In parallel, mock cells are sorted and used as uninfected controls. (B) tSNE dimensionality reduction plot showing the extent of overlap between 3 indicated cell populations from Perth09 experiment clustered based on transcriptional similarity.