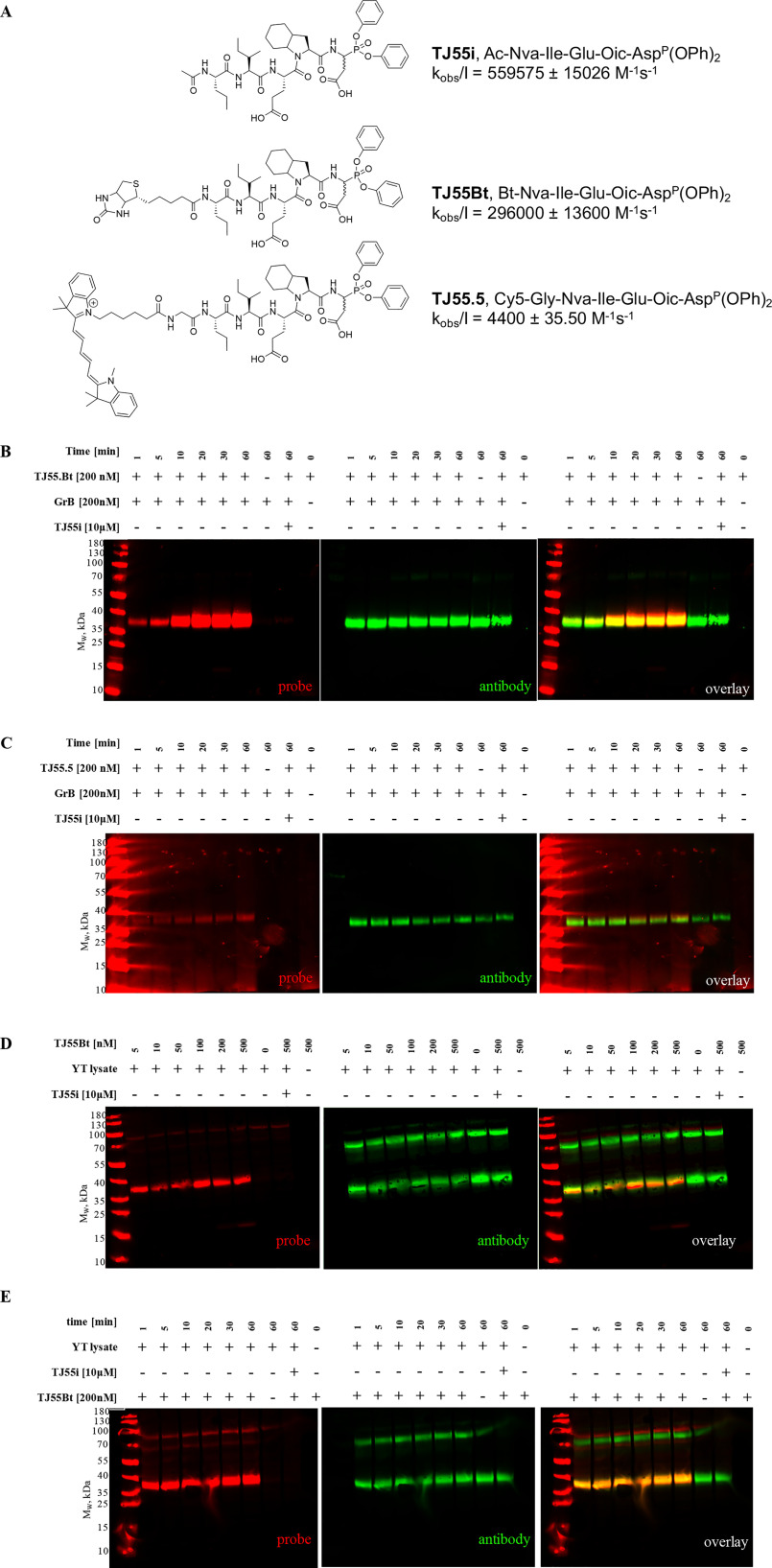
Figure 2.
Specific granzyme B detection in cell lysates. A, covalent GrB inhibitor, biotinylated and fluorescent activity-based probe structures, and kobs/I values for GrB calculated using GraphPad Prism software. Data are reported as the mean ± standard deviation and represent at least 2 independent experiments. B and C, optimization of the recombinant enzyme incubation time with TJ55.Bt (red) (A) or TJ55.5 (red) (B). GrB was incubated with TJ55.Bt or TJ55.5 for the indicated times (only the probe or only the enzyme was used in the controls). Samples then were analyzed by SDS-PAGE, followed by transfer to the membrane and streptavidin conjugate (red) and antibody labeling (green). D and E, YT cell lysates corresponding to 1 × 107 cells/ml were treated with TJ55.Bt (red) for the indicated time (D), or samples were incubated with various concentrations of GrB probe (from 5–500 nm) for 60 min (E). Samples then were analyzed by SDS-PAGE, followed by transfer to the nitrocellulose membrane, streptavidin conjugate labeling (red), and immunoblotting using anti-GrB (green). As a control, lysates were pretreated with a competitive inhibitor prior to probe addition, or the probe and the lysates were run separately. The data reflect at least three separate biological replicates.