Figure 2.
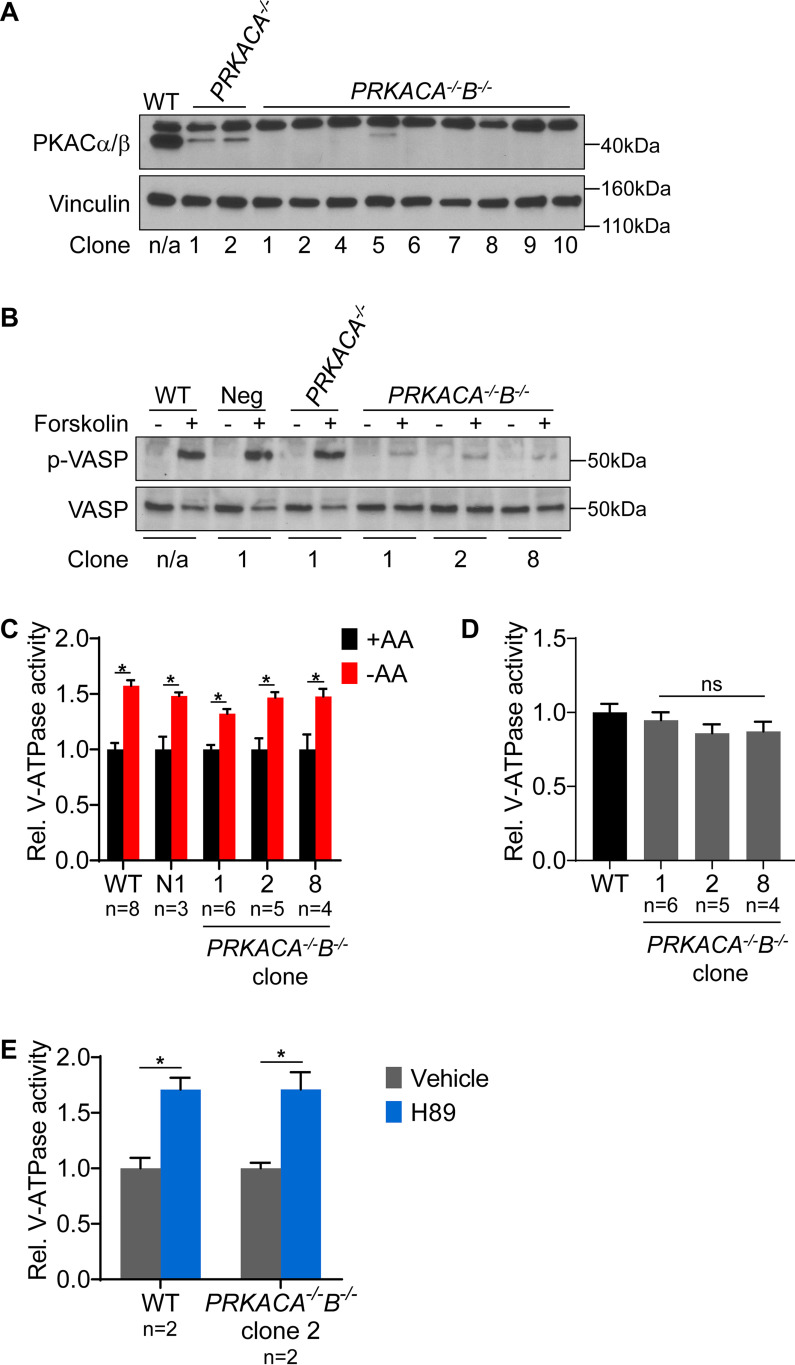
PKA is not required for increased lysosomal V-ATPase activity upon amino acid starvation. A, CRISPR-mediated genome editing was used to disrupt PKA by targeting the PRKACA and PRKACB genes as described in Experimental procedures. Western blotting was performed on lysates from untransfected WT cells (WT), clones targeted for disruption of PKA Cα followed by a nontargeting control (PRKACA−/−), and clones targeted for disruption of PKA Cα followed by the disruption of PKA Cβ (PRKACA−/−B−/−). The antibody recognizes both Cα and Cβ isoforms at the correct molecular weight of 40 kDa. The antibody also produces a nonspecific band at ∼50 kDa. Successful knockout results in loss of the 40-kDa band. Vinculin was used as a loading control. Representative images are shown (n = 2). B, To assess PKA activity, WT, nontarget negative control (Neg), PKA Cα knockout (PRKACA−/−), and PKA Cα/Cβ double knockout (PRKACA−/−B−/−) HEK293T cells were stimulated with DMSO or 50 μm forskolin for 1 h, followed by Western blotting to detect phosphorylation of VASP. Representative images are shown (n = 2). C, WT, nontargeted negative control (Neg), and PKA Cα/Cβ double knockout (PRKACA−/−B−/−) HEK293T cells were incubated with FITC-dextran and then either maintained in amino acids (+AA) or starved (−AA) for 1 h. The rate of V-ATPase–dependent fluorescence quenching in lysosomes was determined as described previously. Values are expressed relative to the unstarved condition for each clone tested. *, p < 0.05; ns, p ≥ 0.05. Error bars represent S.E. D, Lysosomal V-ATPase activity for unstarved PKA Cα/Cβ double knockout clones (PRKACA−/−B−/−) is shown relative to the unstarved WT cells. ns, p ≥ 0.05. Error bars represent S.E. E, The rate of V-ATPase–dependent fluorescence quenching in FITC-dextran-loaded lysosomes was determined for WT and PKA Cα/Cβ double knockout HEK293T cells (PRKACA−/−B−/−) treated for 1 h with either DMSO (vehicle) or 50 μm H89 in the presence of amino acids. Values are expressed relative to the unstarved, vehicle-treated condition. *, p < 0.05; ns, p ≥ 0.05. Error bars represent S.E.