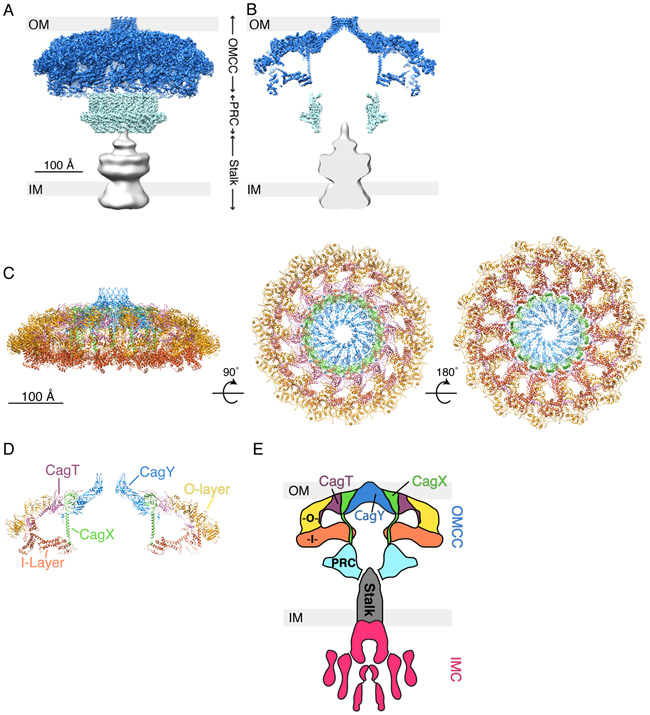
Fig. 3.
3D structure of Cag T4SS particles extracted from H. pylori and determined by single particle cryo-EM. (A) 3D reconstruction of the Cag T4SS with the predicted position of the bacterial outer membrane (OM) and inner membrane (IM) shown (EMD-20023, 20020, and 20021) (16). Outer membrane core complex (OMCC), blue; periplasmic ring complex (PRC), cyan; and stalk, grey. (B) Central axial slice view of the 3D structure in panel A. (C) Secondary structure model of the Cag T4SS OMCC showing the positions of CagT (purple, PDB-6OEE), the C-terminal portion of CagX (green, PDB-6OEG), and the C-terminal portion of CagY (blue, PDB-6ODI), as well as unassigned outer-layer (O-layer, yellow, PDB-6OEF) and inner-layer (I-layer, orange, PDB-6OEH) densities (16). The structure is rotated 90° and then 180° around the X-axis. (D) Central slice view of the structure shown in panel C (left image). (E) Schematic summarizing our current understanding of Cag T4SS architecture, based on cryo-ET and single particle EM studies (13, 14, 16). The Cag T4SS is composed of an OMCC, periplasmic ring complex (PRC, cyan), stalk (grey), and inner membrane complex (IMC, magenta). The positions of CagT (purple) and portions of CagX (green) and CagY (blue) have been mapped into the OMCC. There are outer and inner layer densities (O, yellow and I, orange) that have yet to be assigned a specific T4SS component. The identities and position of T4SS components in the PRC and stalk also have not yet been structurally determined.