Figure 2.
Structure-Function Relationship Between Cardiac Myocytes and Neurons
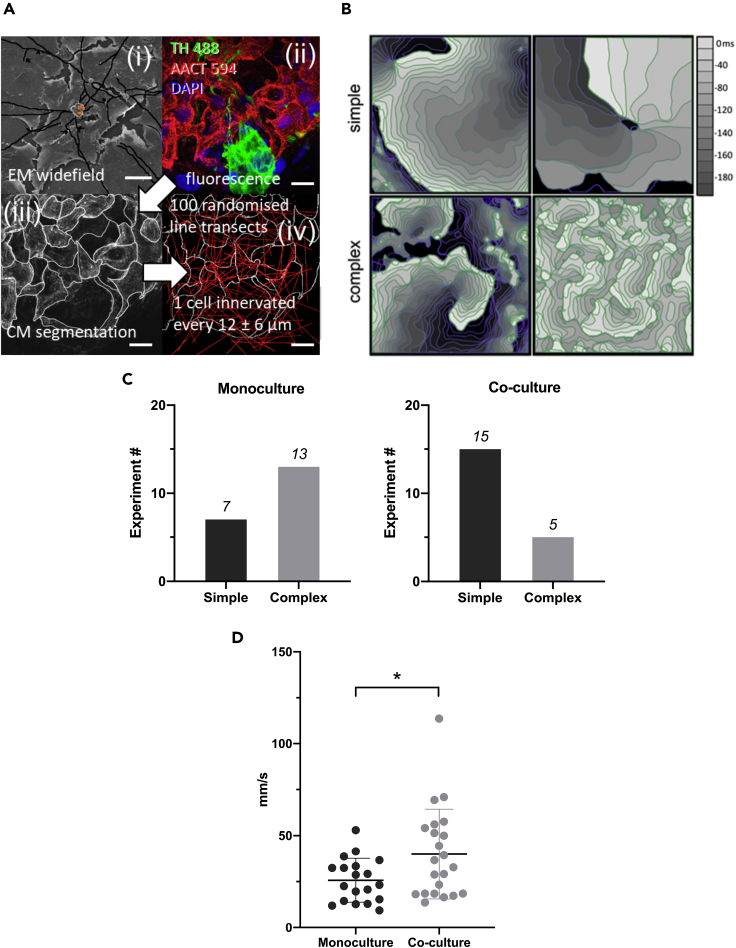
(A) (i) Wide-field scanning electron microscopy image of neuron cell bodies (square orange overlay) showing manual segmentation of dendritic processes traversing the CM monolayer (black lines). See Figure S1 for further images. (ii) Single slice from multichannel confocal stack of CM cells in monolayer (red) around a cell body (green) with nuclei (blue). (iii) Maximum intensity projection of CM channel from (ii) was manually segmented to highlight cell boundaries (white lines). (iv) Randomized linear trajectories were taken through the segmented image to identify the average distance between cell boundaries, indicating the total number of cells that can be innervated by a given length of dendritic process. Scale bars: (i) 50 μm; (ii)–(iv) 20 μm.
(B) Wave dynamics measured by dye-free imaging in Oxford monocultures and co-cultures (see also Figure S5). Isochronal maps of wave dynamics in confluent cardiac-stellate neuron co-cultures display a variety of complex rhythms similar to those seen in intact hearts. Wave dynamics here are classified as simple (top left: targets or top right: single spirals) or complex (bottom left: multiple spiral waves or bottom right: wavelets of activity).
(C) Monocultures display more complex dynamics than co-cultures, which display predominantly simple wavefronts with few wave breaks (p < 0.05, Chi-square).
(D) Comparison of 90-percentile of wave speed for monocultures (25.73 ± 11.88 mm/s, n = 19) and co-cultures 39.96 ± 24.37 (n = 22). Normal distribution of the data was confirmed using the Kolmogorov-Smirnov test, and data were compared using unpaired, two-tailed t-test (∗ indicates p = 0.026). Horizontal bars in D indicate data means ± stdev.
See Figure S13 for wave speeds in SBU cultures.