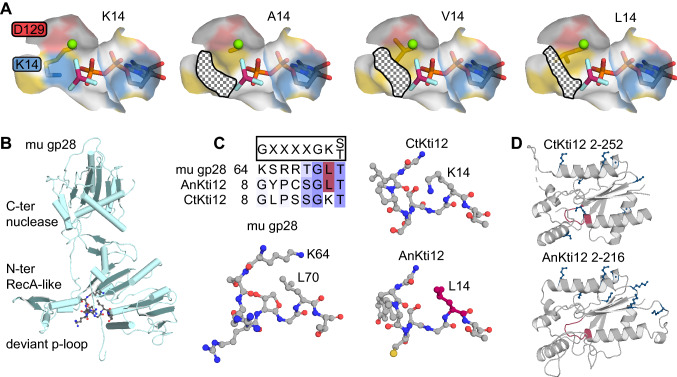
Fig. 4.
Influence of K14L on the nucleotide binding pocket of Kti12 protein. a Hydrophobicity of nucleotide binding pocket of Kti12 mutants. PDB ID: 6QP0. Nucleotide binding pocket consists of positively charged (blue), negatively charged (red) and hydrophobic (yellow) residues. Only surface of the residues within 5 Å from the phosphor atoms of ADP is shown. Kti12 is shown bound to ADP (standard coloring), AlF3 (cyan and pink) and magnesium (green). K14A mutation creates a relatively big gap, leaving the hydrophobic core unprotected. Subsequent K14V and K14L substitutions gradually repair the breach. b A homology-based model of gp28 terminase (based on PDB ID 3EZK) from mu phage with previously described deviant Walker-A motif shown in ball and stick representation. c Multiple sequence alignment and structural comparison of Walker A motifs in CtKti12, AnKti12 (based on 6QP0) and mu phage gp28. Shades of violet indicate conservation score. Please note that AnKti12 Walker A motif does not have additional lysine which is present in case of mu gp28, but has L14 residue (pink) which corresponds to L70 residue of mu gp28. d An overview of lysines (dark blue) present in the ATPase domain of CtKti12 (PDB ID 6QP0, top) and AnKti12 (based on PDB ID 6QP0; bottom). Walker A motif is highlighted in pink